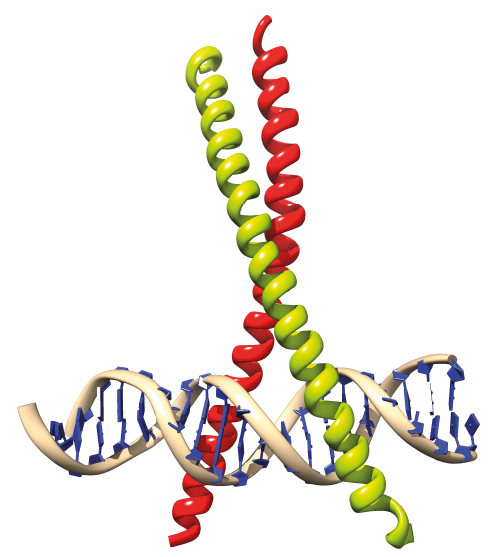
|
| AC023509.3 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
100 perc ID - in vitro |
|
Identical bZIP DBD to ATF7. Some isoforms also contain a single C2H2 ZF. This is known to facilitate protein interactions in ATF7 (see ATF7 Uniprot page), so AC023509.3 is only classified here as a bZIP protein |
|
| APC2 |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
Based on the alignments, the protein is only weakly related to the other bZIP proteins and lacks the basic region required for DNA binding |
|
| ATF1 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
In vivo/Misc source |
Only known motifs are from Transfac or HocoMoco - origin is uncertain |
|
|
| ATF2 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| ATF3 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| ATF4 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| ATF5 |
bZIP |
Yes |
Known motif |
2 Obligate heteromer |
In vivo/Misc source |
Only known motifs are from Transfac or HocoMoco - origin is uncertain |
Transfac motifs dont correspond to canonical bZIP binding sites. Annotated as obligate heteromer based on peptide array studies (PMID:12805554). |
|
| ATF6 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| ATF6B |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| ATF7 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
Some isoforms also contain a single C2H2 ZF. This is known to facilitate protein interactions (see Uniprot page), so ATF7 is only classified here as a bZIP protein |
|
| BACH1 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
In vivo/Misc source |
Only known motifs are from Transfac or HocoMoco - origin is uncertain |
|
|
| BACH2 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
Based on Newman et al 2003 (PMID: 12805554), the protein has strong preference for forming heterodimers with MAFG and MAFK over homo-dimerisation. The Homer ChIP-seq motif appears to be a MAF-BACH2 heterodimer. |
|
| BATF |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
Prefers to heterodimerize, but it has an HT-SELEX (homodimer) motif. Its homodimerization Kd is 184nM and its best heterodimer is with CEBPG (<1nM) (PMID: 23661758). |
|
| BATF2 |
bZIP |
Yes |
Likely to be sequence specific TF |
2 Obligate heteromer |
No motif |
|
Part of AP-1 complex - there is no evidence it can bind on its own as a homodimer. |
|
| BATF3 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
Transfac motif is dubious. |
|
| BEND4 |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
Lacks the basic region of the bZIP domain. |
|
| CCDC3 |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
bZIP domain is truncated and lacks the DNA-contacting residues. |
|
| CCDC83 |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
bZIP domains are truncated, such that they are likely just coiled-coiled regions with no DNA-binding ability. |
|
| CEBPA |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
100 perc ID - in vitro |
|
|
|
| CEBPB |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| CEBPD |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
CEBPD binds as both a homodimer and as a heterdimer with other C/EBP TFs (PMID: 1884998; PMID: 12805554). |
|
| CEBPE |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
Also forms heterodimers. |
|
| CEBPG |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| CREB1 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| CREB3 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| CREB3L1 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| CREB3L2 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
100 perc ID - in vitro |
|
|
|
| CREB3L3 |
bZIP |
Yes |
Likely to be sequence specific TF |
1 Monomer or homomultimer |
No motif |
|
Gel-shift and reporter experiments demonstrate that CREB3L3 is a TF (PMID: 11353085). |
|
| CREB3L4 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| CREB5 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| CREBL2 |
bZIP |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| CREBRF |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
Tested on HT-SELEX and did not yield a motif. Binds to CREB3 and represses the unfolded protein response (PMID: 18391022). |
|
| CREBZF |
bZIP |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| CREM |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| DBP |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| DDIT3 |
bZIP |
Yes |
Known motif |
2 Obligate heteromer |
In vivo/Misc source |
Only known motifs are from Transfac or HocoMoco - origin is uncertain |
Has strong preference for forming heterodimers with BATF; DBP; HLF; CEBP[ABCD] over homodimerization (PMID: 12805554). |
|
| FOS |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| FOSB |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| FOSL1 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| FOSL2 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
Also forms heterodimers. |
|
| GULP1 |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
Based on the alignments, the protein is only weakly related to the other bZIP proteins and lacks the basic region required for DNA binding |
|
| HLF |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| HOOK2 |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
Based on the alignments, the protein is only weakly related to the other bZIP proteins and lacks the basic region required for DNA binding |
|
| IQGAP1 |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
bZIP domain in the protein is just a fragment. |
|
| JDP2 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| JUN |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| JUNB |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
Prefers forming heterodimers with FOS; FOSB; FOSL1 and FOSL2 over homodimers (PMID:12805554); but, clearly can bind DNA specifically in vitro. |
|
| JUND |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| KIF15 |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
Has a bZIP like fragment that lacks the basic region required for DNA binding and a STE-domain that is classified as a potential DBD in CIS-BP. It is a kinesin operating in the microtubule system (PMID: 24419385) |
|
| KRT13 |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
Based on the alignments, the protein is only weakly related to the other bZIP proteins and lacks the basic region required for DNA binding |
|
| MAF |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| MAFA |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| MAFB |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
100 perc ID - in vitro |
|
PDB:2WTY is a homodimer crystallised with TAATTGCTGACTCAGCAAAT sequence |
|
| MAFF |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| MAFG |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| MAFK |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| MORC3 |
bZIP |
No |
ssDNA/RNA binding |
4 Not a DNA binding protein |
No motif |
|
Lacks a canonical bZIP DBD but: regulates TP53 activity (PMID: 17332504]), binds RNA in vitro (PMID: 11927593), and may be required for influenza A transcription during viral infection (PMID: 26202233). |
|
| NDC80 |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
Based on the alignments, the protein is only weakly related to the other bZIP proteins and lacks the basic region required for DNA binding |
|
| NFE2 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| NFE2L1 |
bZIP |
Yes |
Known motif |
2 Obligate heteromer |
In vivo/Misc source |
Only known motifs are from Transfac or HocoMoco - origin is uncertain |
Tested on HT-SELEX and PBM. Neither yielded a motif. Likely an obligate heteromer (PMID: 23661758). |
|
| NFE2L2 |
bZIP |
Yes |
Known motif |
2 Obligate heteromer |
In vivo/Misc source |
Only known motifs are from Transfac or HocoMoco - origin is uncertain |
Has been tested by both PBM and HT-SELEX. Neither yielded a motif. Likely obligate heteromer. |
|
| NFE2L3 |
bZIP |
Yes |
Known motif |
2 Obligate heteromer |
In vivo/Misc source |
Only known motifs are from Transfac or HocoMoco - origin is uncertain |
Tested on HT-SELEX and PBM. Neither yielded a motif. Likely an obligate heteromer (PMID: 23661758). Strong preference for forming heterodimers with MAFG and MAFK over homodimers (PMID: 12805554). |
|
| NFIL3 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| NRBF2 |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
Based on the alignments, the protein is only weakly related to the other bZIP proteins and lacks the basic region required for DNA binding |
|
| NRL |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| PPP1R21 |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
Protein lacks the basic region these proteins use to contact the DNA |
|
| RAB11FIP4 |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
Likely false-positive - appears to function primarily in the cytoplasm (PMID: 12470645). |
|
| RNF219 |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
Not a proper bZIP; contains only a small leucine zipper region but lacks the DNA-interfacing basic part. |
|
| SCOC |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
Not a proper bZIP; contains only a small leucine zipper region but lacks the DNA-interfacing basic part. |
|
| TEF |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| TRAK2 |
bZIP |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
The bZIP domain is only a partial 40AA sequence that will not bind DNA. |
|
| XBP1 |
bZIP |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|