| Source |
Annotation |
Motif |
Evidence |
| HT-SELEX | Jolma2013 |  | Direct |
| HT-SELEX | Yin2017 |  | Direct |
| Methyl-HT-SELEX | Yin2017 | 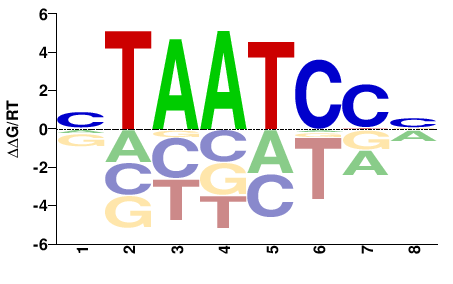 | Direct |
| PBM | Berger08 | 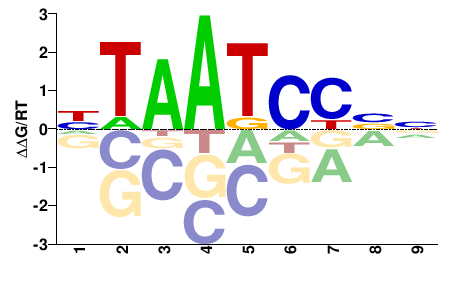 | Inferred - Dmbx1 (100% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 | 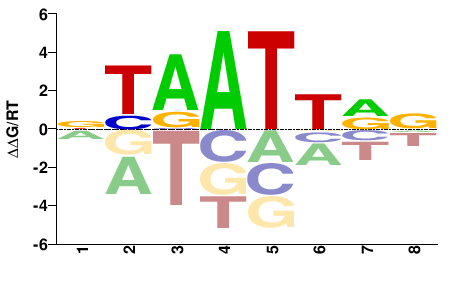 | Inferred - ALX4 (75% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - ALX4 (75% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 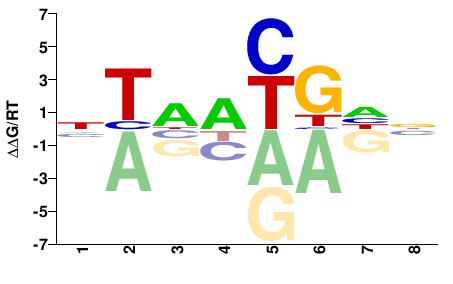 | Inferred - ALX4 (75% AA Identity, Homo sapiens) |
| PBM | Berger08 |  | Inferred - Alx4 (75% AA Identity, Mus musculus) |
| Transfac | Transfac | License required | Inferred - ALX4 (75% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - ALX1 (73% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - ALX1 (73% AA Identity, Homo sapiens) |
| PBM | Berger08 | 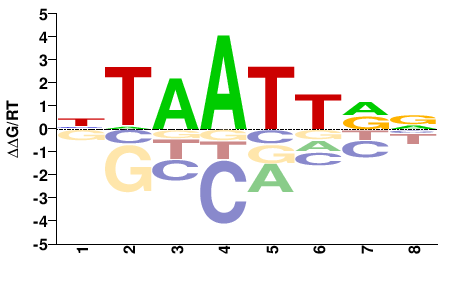 | Inferred - Alx1 (73% AA Identity, Mus musculus) |
| PBM | Berger08 | 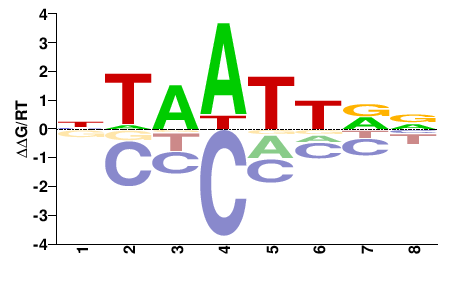 | Inferred - Alx1 (73% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 | 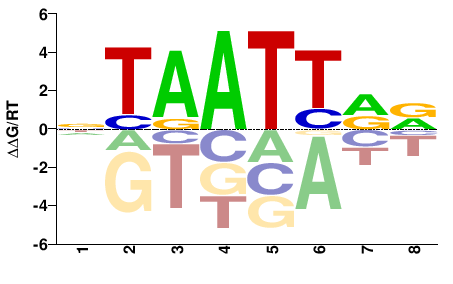 | Inferred - DRGX (73% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 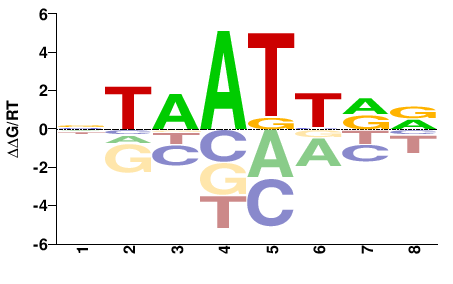 | Inferred - DRGX (73% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - ALX1 (73% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - Drgx (73% AA Identity, Rattus norvegicus) |
| Misc | HocoMoco |  | Inferred - ALX1 (73% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 | 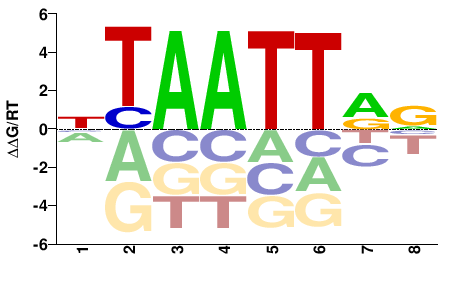 | Inferred - ALX3 (71% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 | 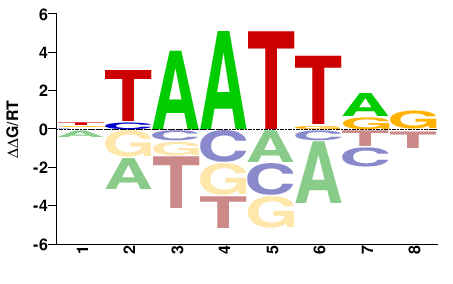 | Inferred - ALX3 (71% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 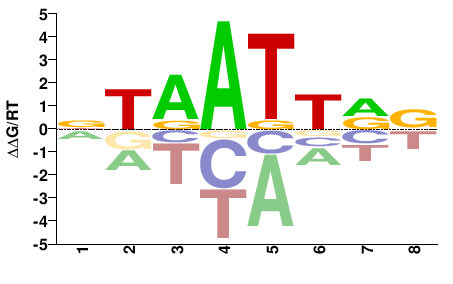 | Inferred - ALX3 (71% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 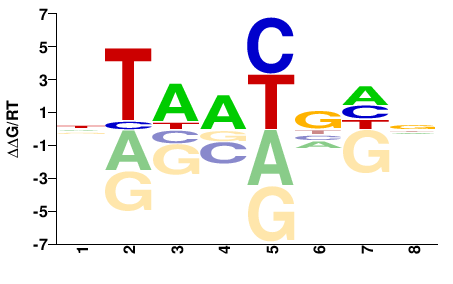 | Inferred - ALX3 (71% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 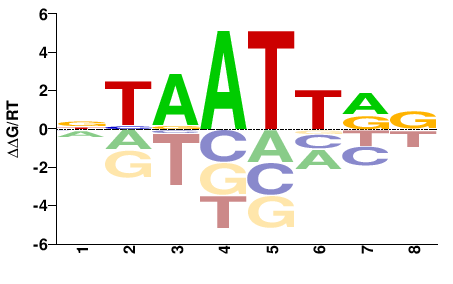 | Inferred - ALX3 (71% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 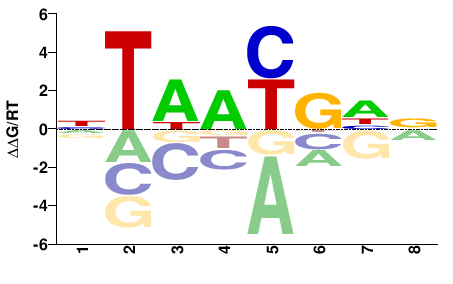 | Inferred - ALX3 (71% AA Identity, Homo sapiens) |
| PBM | Berger08 | 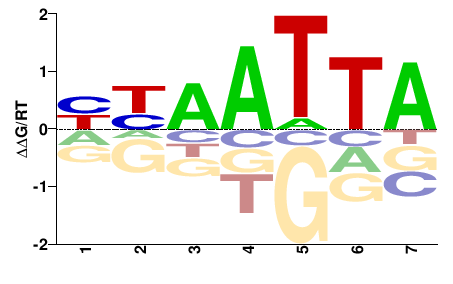 | Inferred - Alx3 (71% AA Identity, Mus musculus) |
| PBM | Barrera2016 | 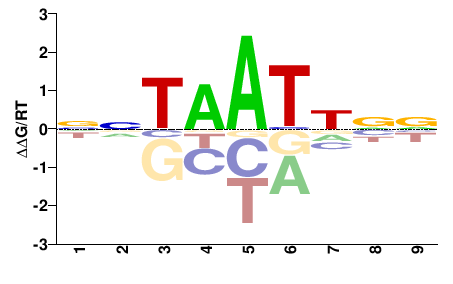 | Inferred - ARX (71% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - ARX (71% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 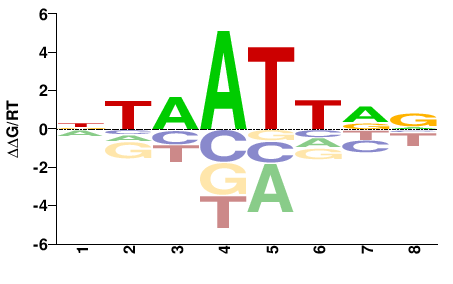 | Inferred - ARX (71% AA Identity, Homo sapiens) |
| PBM | Berger08 | 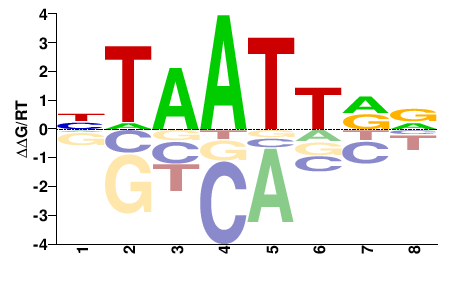 | Inferred - Arx (71% AA Identity, Mus musculus) |
| HT-SELEX | Nitta2015 |  | Inferred - CG2808 (71% AA Identity, Drosophila melanogaster) |
| Transfac | Transfac | License required | Inferred - Arx (71% AA Identity, Rattus norvegicus) |
| PBM | Barrera2016 |  | Inferred - ARX (70% AA Identity, Homo sapiens) |
| PBM | Barrera2016 | 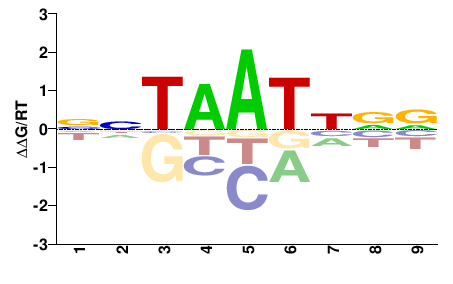 | Inferred - ARX (70% AA Identity, Homo sapiens) |
| PBM | Barrera2016 |  | Inferred - ARX (70% AA Identity, Homo sapiens) |
| PBM | Barrera2016 | 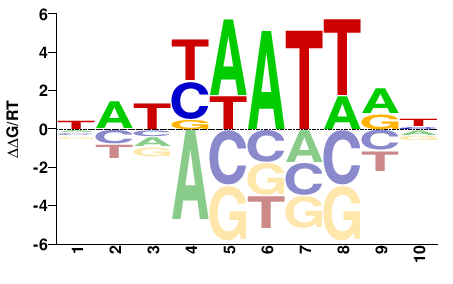 | Inferred - ARX (70% AA Identity, Homo sapiens) |
| PBM | Barrera2016 | 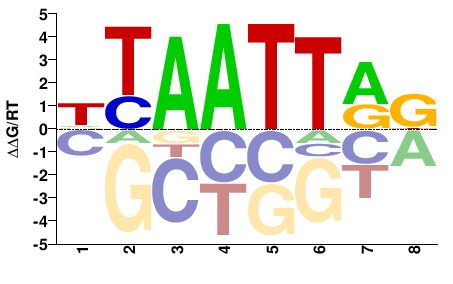 | Inferred - ARX (70% AA Identity, Homo sapiens) |
| SMiLE-seq | Isakova2017 |  | Inferred - Gsc (70% AA Identity, Drosophila melanogaster) |
| B1H | JASPAR | 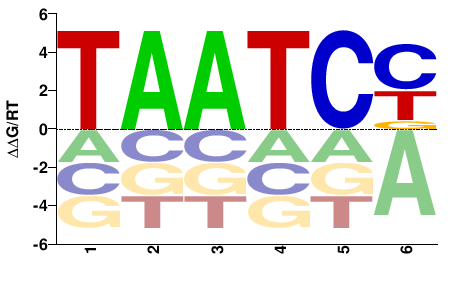 | Inferred - Gsc (70% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 | 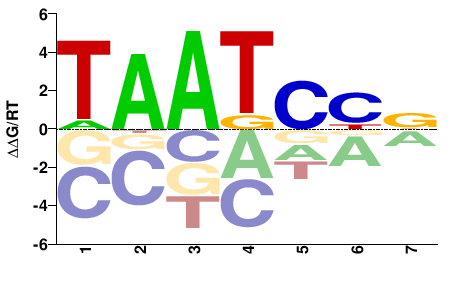 | Inferred - Gsc (70% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 | 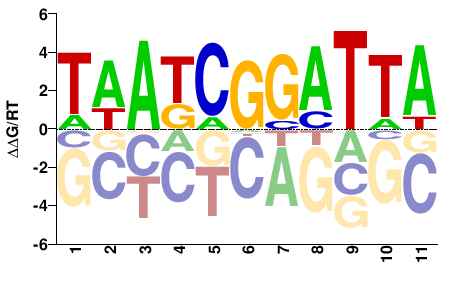 | Inferred - Gsc (70% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 | 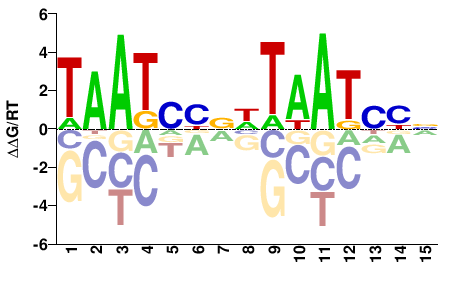 | Inferred - Gsc (70% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 |  | Inferred - Gsc (70% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 | 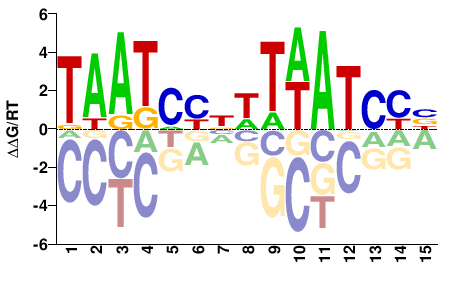 | Inferred - Gsc (70% AA Identity, Drosophila melanogaster) |
| PBM | Zoo_01 | 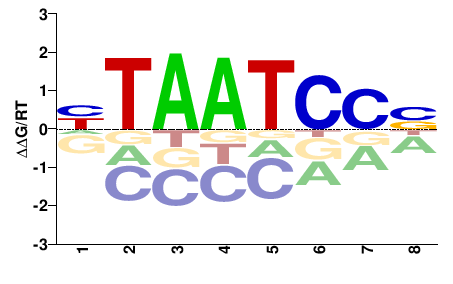 | Inferred - Gsc (70% AA Identity, Drosophila melanogaster) |
| B1H | JASPAR |  | Inferred - al (68% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 | 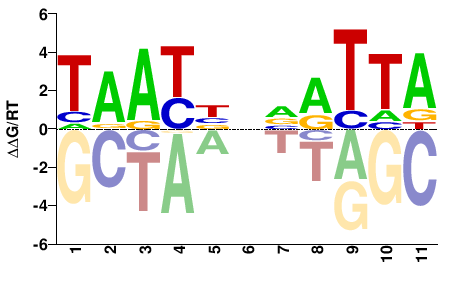 | Inferred - al (68% AA Identity, Drosophila melanogaster) |
| B1H | JASPAR |  | Inferred - CG11294 (68% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 |  | Inferred - CG11294 (68% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 |  | Inferred - CG11294 (68% AA Identity, Drosophila melanogaster) |
| B1H | JASPAR |  | Inferred - hbn (68% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 |  | Inferred - hbn (68% AA Identity, Drosophila melanogaster) |
| PBM | Barrera2016 |  | Inferred - VSX1 (68% AA Identity, Homo sapiens) |
| PBM | Barrera2016 | 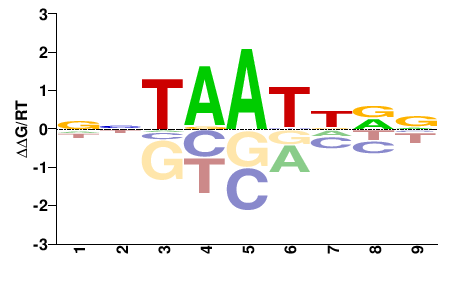 | Inferred - VSX1 (68% AA Identity, Homo sapiens) |
| PBM | Berger08 | 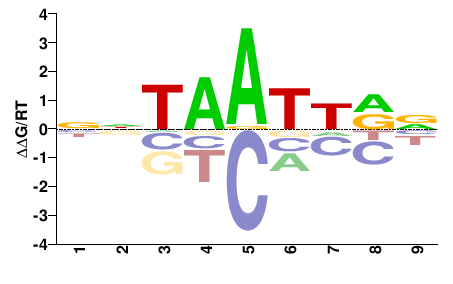 | Inferred - Vsx1 (68% AA Identity, Mus musculus) |
| PBM | Barrera2016 | 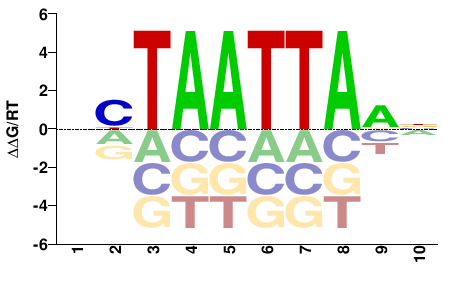 | Inferred - VSX2 (68% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - VSX2 (68% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - VSX2 (68% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - VSX2 (68% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - VSX2 (68% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 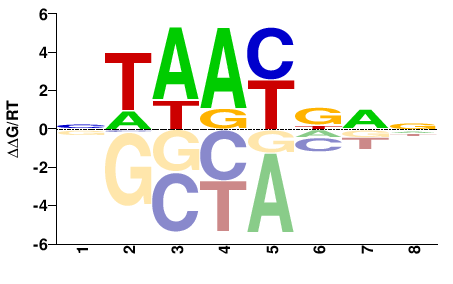 | Inferred - VSX2 (68% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - VSX2 (68% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - VSX2 (68% AA Identity, Homo sapiens) |
| Misc | HocoMoco | 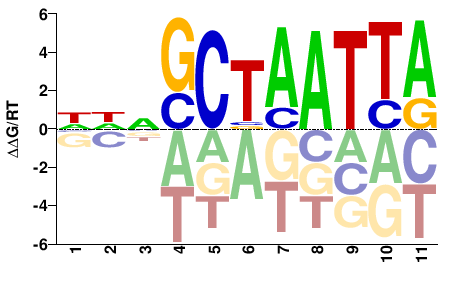 | Inferred - VSX2 (68% AA Identity, Homo sapiens) |
| PBM | Barrera2016 | 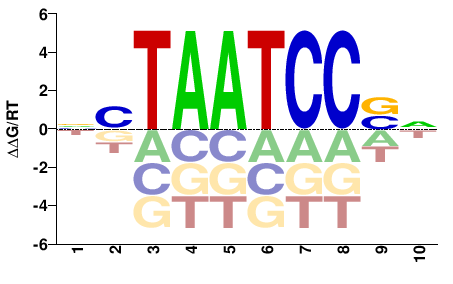 | Inferred - CRX (66% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - CRX (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 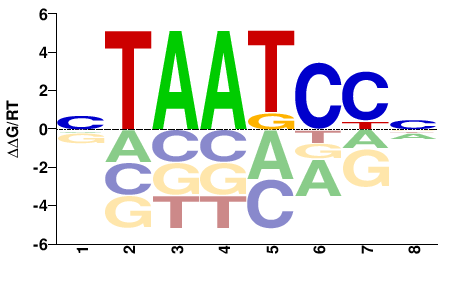 | Inferred - CRX (66% AA Identity, Homo sapiens) |
| PBM | Berger08 | 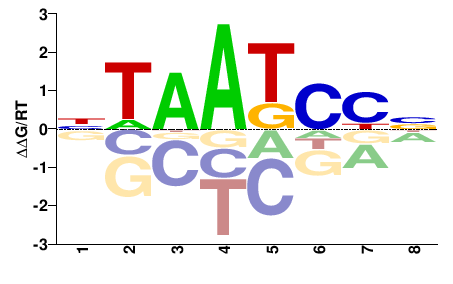 | Inferred - Crx (66% AA Identity, Mus musculus) |
| B1H | JASPAR |  | Inferred - Rx (66% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 | 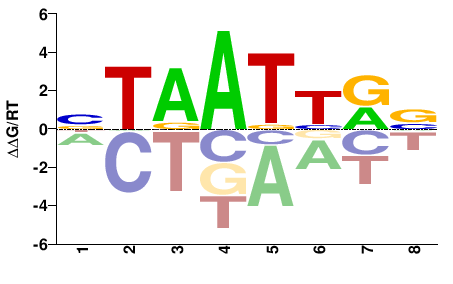 | Inferred - Rx (66% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 | 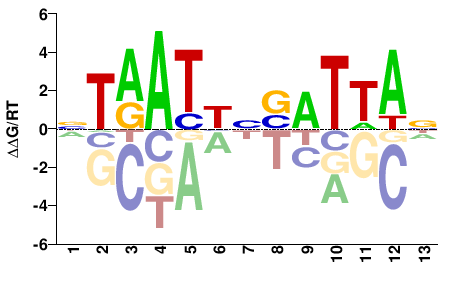 | Inferred - Rx (66% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Yin2017 | 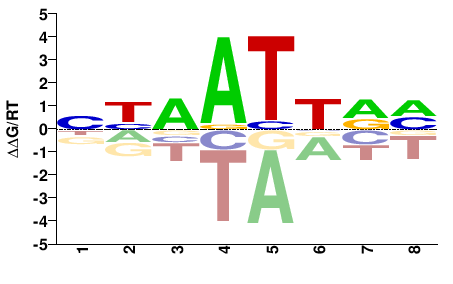 | Inferred - UNCX (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 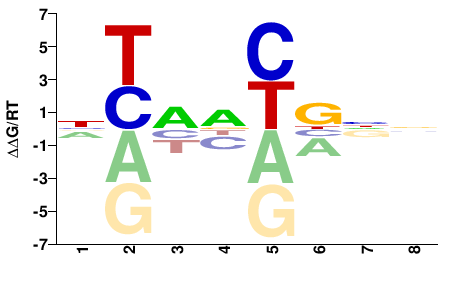 | Inferred - UNCX (66% AA Identity, Homo sapiens) |
| PBM | Berger08 | 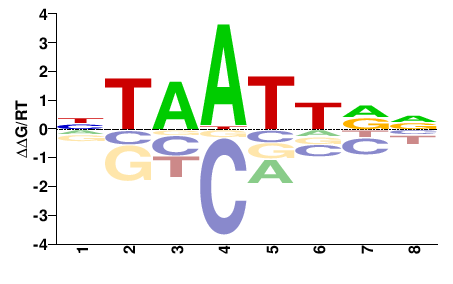 | Inferred - Uncx (66% AA Identity, Mus musculus) |
| PBM | Zoo_01 |  | Inferred - uncx4.1 (66% AA Identity, Tetraodon nigroviridis) |
| PBM | Barrera2016 | 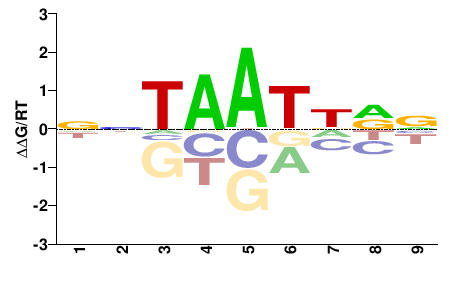 | Inferred - VSX1 (66% AA Identity, Homo sapiens) |
| PBM | Zoo_01 | 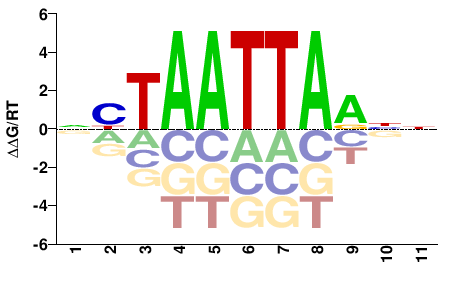 | Inferred - vsx2 (66% AA Identity, Danio rerio) |
| Transfac | Transfac | License required | Inferred - CRX (66% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - CRX (66% AA Identity, Homo sapiens) |
| ChIP-seq | JASPAR |  | Inferred - Crx (66% AA Identity, Mus musculus) |
| Misc | HocoMoco |  | Inferred - CRX (66% AA Identity, Homo sapiens) |
| Misc | HOMER | 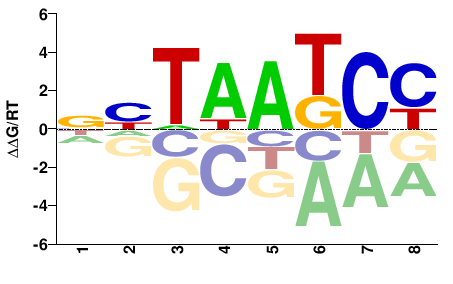 | Inferred - Crx (66% AA Identity, Mus musculus) |
| PBM | BusserDevelopment2012 | 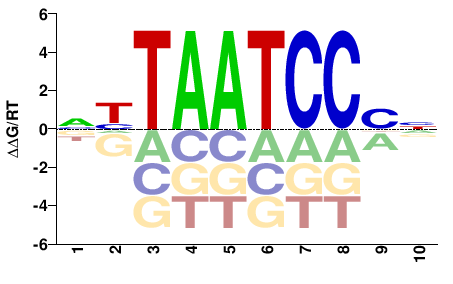 | Inferred - Ptx1 (65% AA Identity, Drosophila melanogaster) |
| B1H | JASPAR | 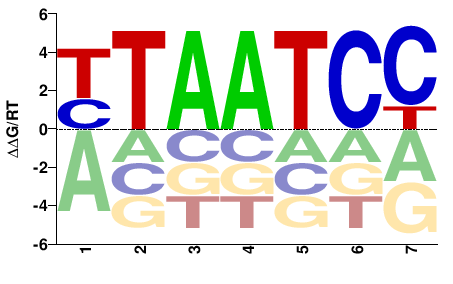 | Inferred - Ptx1 (65% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 | 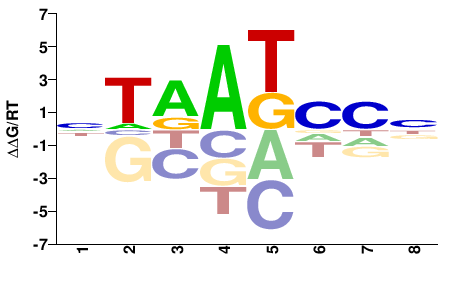 | Inferred - Ptx1 (65% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 |  | Inferred - Ptx1 (65% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 | 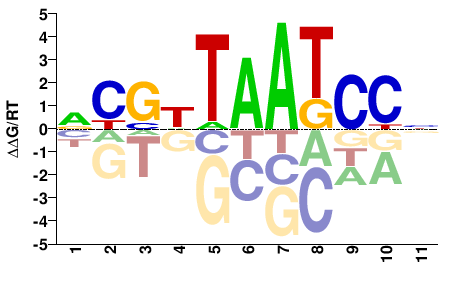 | Inferred - Ptx1 (65% AA Identity, Drosophila melanogaster) |
| PBM | Zoo_01 |  | Inferred - Ptx1 (65% AA Identity, Drosophila melanogaster) |