| Source |
Annotation |
Motif |
Evidence |
| HT-SELEX | Jolma2013 |  | Direct |
| HT-SELEX | Yin2017 |  | Direct |
| HT-SELEX | Yin2017 |  | Direct |
| Methyl-HT-SELEX | Yin2017 | 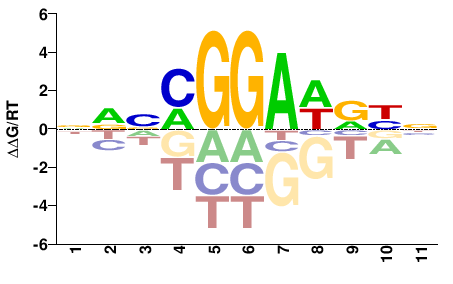 | Direct |
| ChIP-seq | ENCODE |  | Direct |
| ChIP-seq | ENCODE | 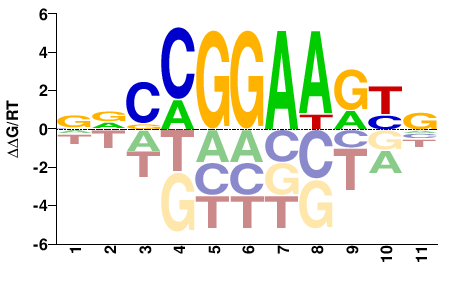 | Direct |
| ChIP-seq | JASPAR |  | Direct |
| Transfac | Transfac | License required | Direct |
| Transfac | Transfac | License required | Direct |
| Misc | HocoMoco |  | Direct |
| PBM | Wei10 | 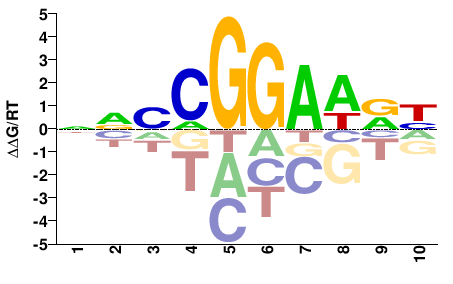 | Inferred - Elk4 (95% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 |  | Inferred - ELK3 (82% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - ELK3 (82% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 | 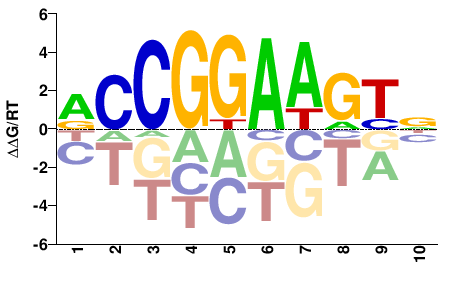 | Inferred - ELK3 (82% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - ELK3 (82% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - ELK3 (82% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - ELK3 (82% AA Identity, Homo sapiens) |
| PBM | Zoo_01 |  | Inferred - elk4 (82% AA Identity, Tetraodon nigroviridis) |
| Transfac | Transfac | License required | Inferred - ELK3 (82% AA Identity, Homo sapiens) |
| Misc | HocoMoco |  | Inferred - ELK3 (82% AA Identity, Homo sapiens) |
| PBM | Wei10 | 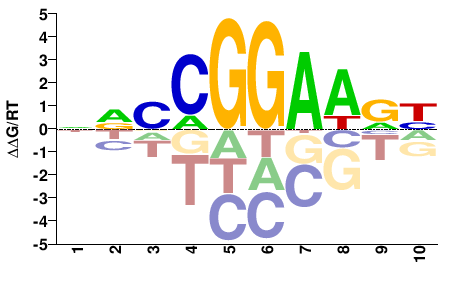 | Inferred - Elk3 (81% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 |  | Inferred - ELK1 (79% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - ELK1 (79% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - ELK1 (79% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 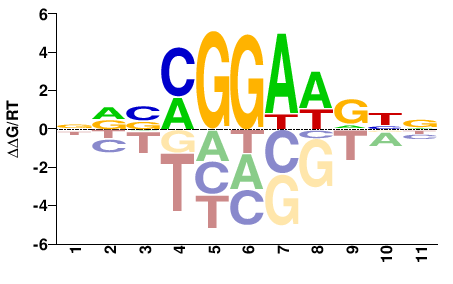 | Inferred - ELK1 (79% AA Identity, Homo sapiens) |
| PBM | Wei10 | 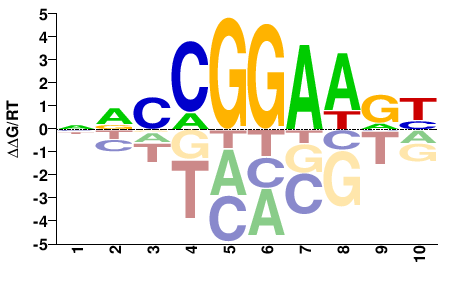 | Inferred - Elk1 (79% AA Identity, Mus musculus) |
| SELEX | JASPAR |  | Inferred - ELK1 (79% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - ELK1 (79% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - ELK1 (79% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - ELK1 (79% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - ELK1 (79% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - ELK1 (79% AA Identity, Homo sapiens) |
| Misc | HocoMoco | 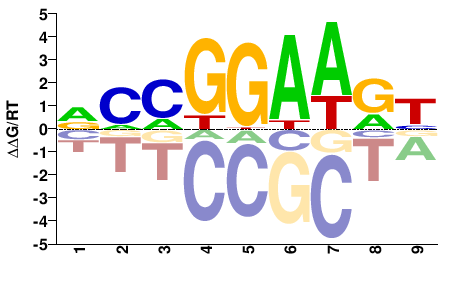 | Inferred - ELK1 (79% AA Identity, Homo sapiens) |
| Misc | HOMER | 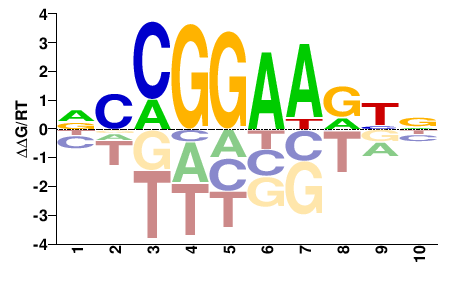 | Inferred - ELK1 (79% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 | 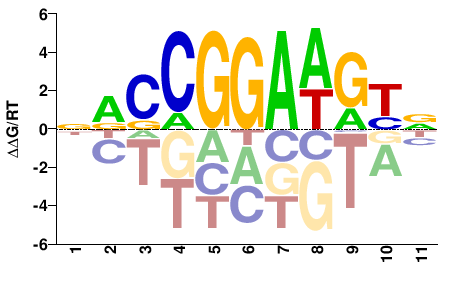 | Inferred - ETV4 (61% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - ETV4 (61% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - ETV4 (61% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 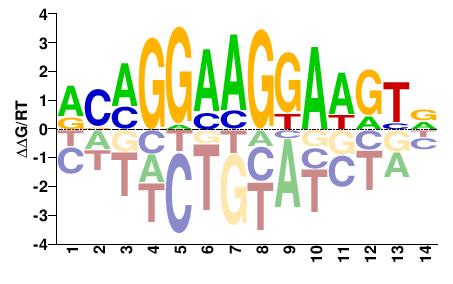 | Inferred - ETV4 (61% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - ETV4 (61% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 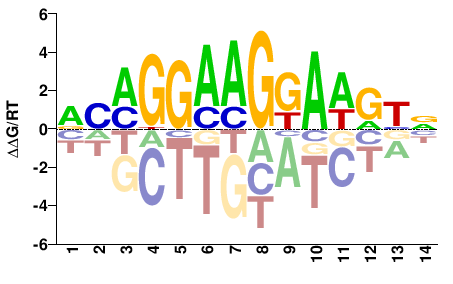 | Inferred - ETV4 (61% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - ETV4 (61% AA Identity, Homo sapiens) |
| PBM | Wei10 | 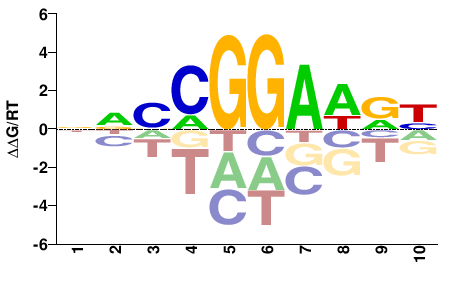 | Inferred - Etv4 (61% AA Identity, Mus musculus) |
| Transfac | Transfac | License required | Inferred - ETV4 (61% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - ETV4 (61% AA Identity, Homo sapiens) |
| Misc | HocoMoco | 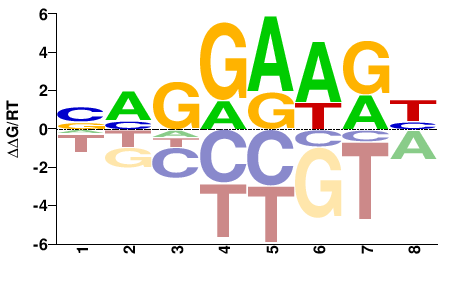 | Inferred - ETV4 (61% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - ETV1 (60% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - ETV1 (60% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 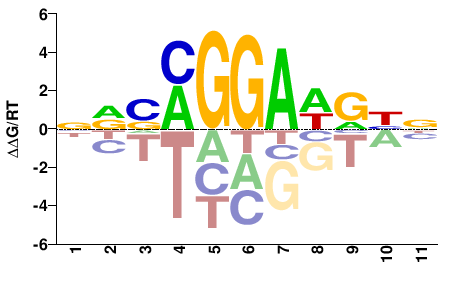 | Inferred - ETV1 (60% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - ETV1 (60% AA Identity, Homo sapiens) |
| PBM | Wei10 | 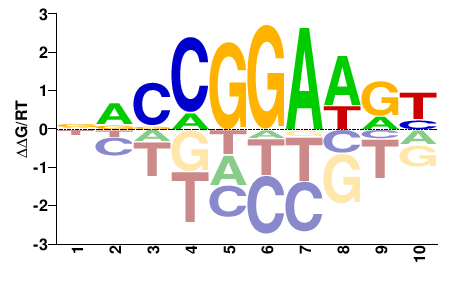 | Inferred - Etv1 (60% AA Identity, Mus musculus) |
| PBM | Wei10 | 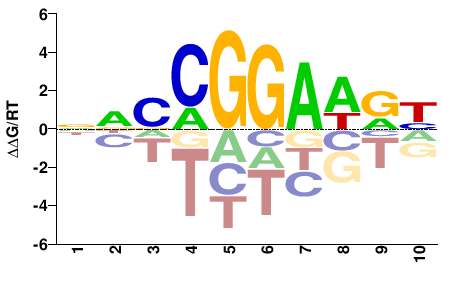 | Inferred - Etv1 (60% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 |  | Inferred - ETV5 (60% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 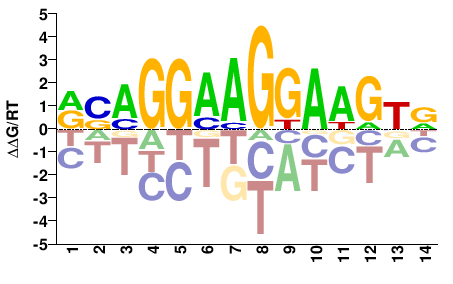 | Inferred - ETV5 (60% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - ETV5 (60% AA Identity, Homo sapiens) |
| PBM | Wei10 | 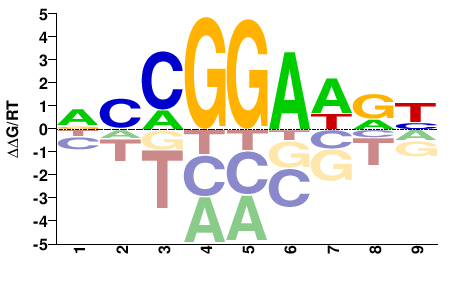 | Inferred - Etv5 (60% AA Identity, Mus musculus) |
| Misc | HOMER |  | Inferred - ETV1 (60% AA Identity, Homo sapiens) |
| Misc | HocoMoco | 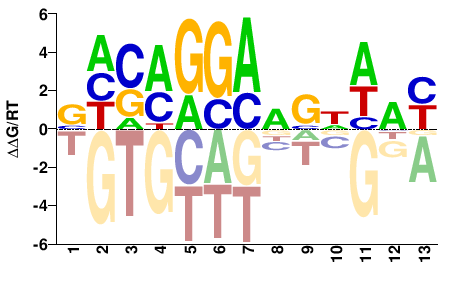 | Inferred - ETV5 (60% AA Identity, Homo sapiens) |
| SMiLE-seq | Isakova2017 |  | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| PBM | Badis09 |  | Inferred - Gabpa (59% AA Identity, Mus musculus) |
| PBM | Wei10 |  | Inferred - Gabpa (59% AA Identity, Mus musculus) |
| HT-SELEX | Nitta2015 |  | Inferred - pnt (59% AA Identity, Drosophila melanogaster) |
| ChIP-seq | ENCODE | 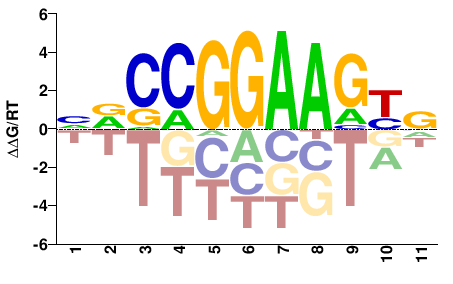 | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| ChIP-seq | ENCODE | 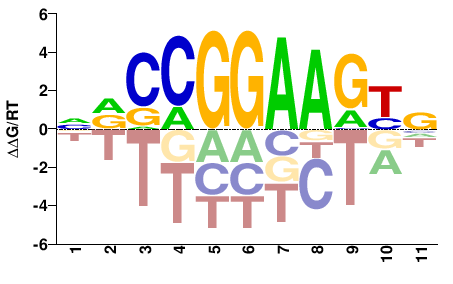 | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| ChIP-seq | ENCODE | 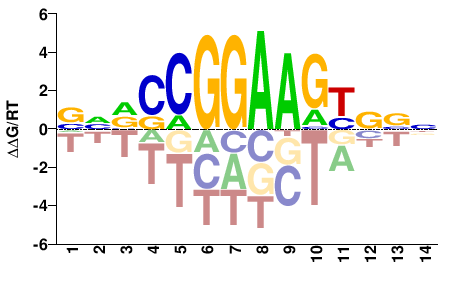 | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| ChIP-seq | ENCODE | 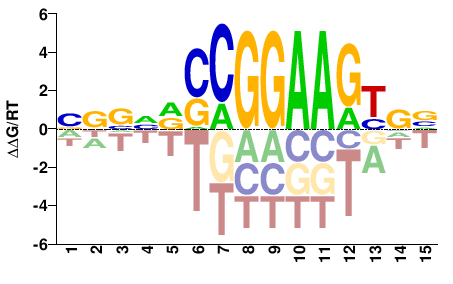 | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| ChIP-seq | ENCODE | 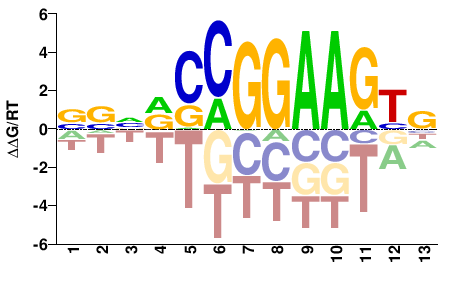 | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| ChIP-seq | JASPAR |  | Inferred - Gabpa (59% AA Identity, Mus musculus) |
| Misc | HocoMoco | 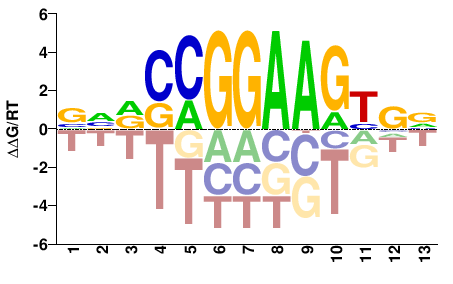 | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| Misc | HOMER |  | Inferred - GABPA (59% AA Identity, Homo sapiens) |
| PBM | Wei10 | 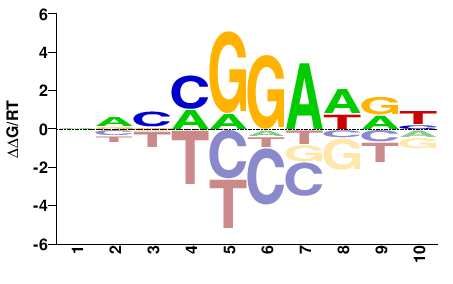 | Inferred - Ets1 (58% AA Identity, Mus musculus) |
| Transfac | Transfac | License required | Inferred - ETS1B_CHICK (58% AA Identity, Gallus gallus) |
| HT-SELEX | Nitta2015 |  | Inferred - Ets96B (55% AA Identity, Drosophila melanogaster) |
| PBM | Wei10 | 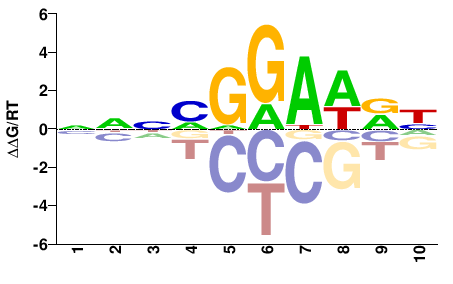 | Inferred - XP_911724.4 (55% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 |  | Inferred - ETV3 (54% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - ETV3 (54% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - ETV3 (54% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 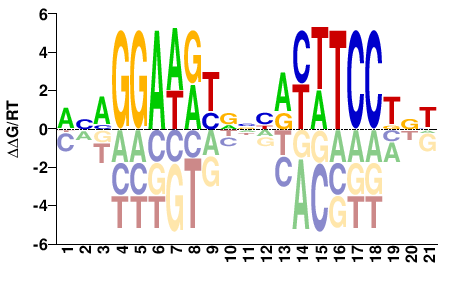 | Inferred - ETV3 (54% AA Identity, Homo sapiens) |
| PBM | Wei10 | 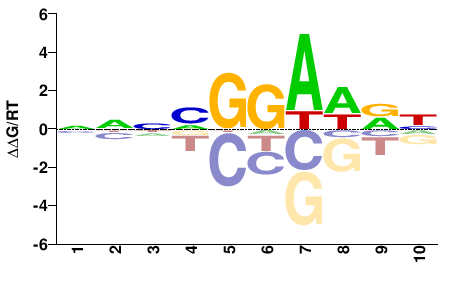 | Inferred - Etv3 (54% AA Identity, Mus musculus) |
| HT-SELEX | Nitta2015 |  | Inferred - Ets65A (53% AA Identity, Drosophila melanogaster) |