| Source |
Annotation |
Motif |
Evidence |
| HT-SELEX | Jolma2013 | 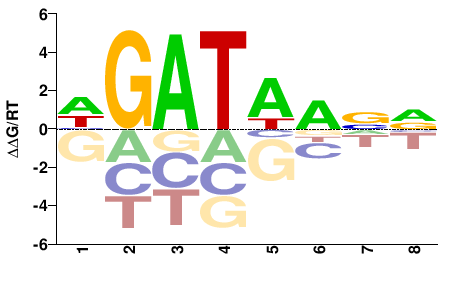 | Direct |
| HT-SELEX | Yin2017 |  | Direct |
| HT-SELEX | Yin2017 |  | Direct |
| HT-SELEX | Yin2017 |  | Direct |
| HT-SELEX | Yin2017 |  | Direct |
| Methyl-HT-SELEX | Yin2017 |  | Direct |
| Methyl-HT-SELEX | Yin2017 |  | Direct |
| Methyl-HT-SELEX | Yin2017 | 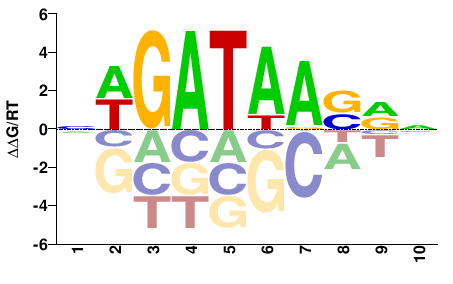 | Direct |
| Methyl-HT-SELEX | Yin2017 |  | Direct |
| Misc | HocoMoco | 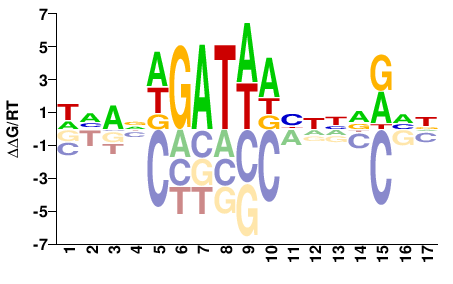 | Direct |
| PBM | Badis09 | 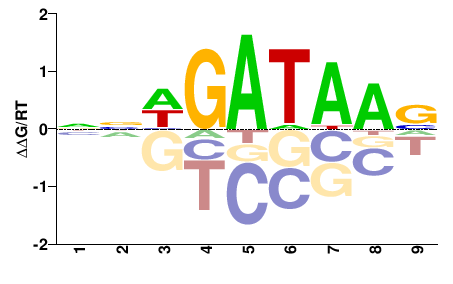 | Inferred - Gata5 (95% AA Identity, Mus musculus) |
| Transfac | Transfac | License required | Inferred - Gata5 (95% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 | 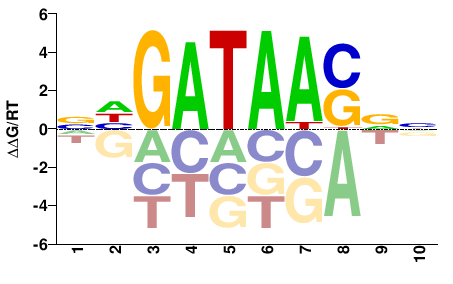 | Inferred - GATA4 (91% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - GATA4 (91% AA Identity, Homo sapiens) |
| PBM | DREAM_contest | 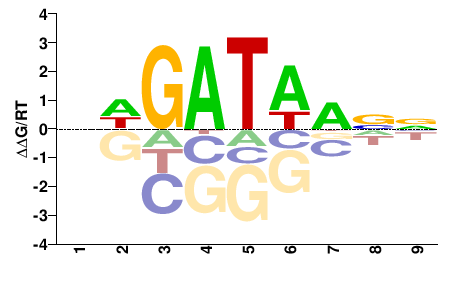 | Inferred - Gata4 (91% AA Identity, Mus musculus) |
| Transfac | Transfac | License required | Inferred - GATA4 (91% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA4 (91% AA Identity, Homo sapiens) |
| ChIP-seq | JASPAR |  | Inferred - Gata4 (91% AA Identity, Mus musculus) |
| Misc | HocoMoco | 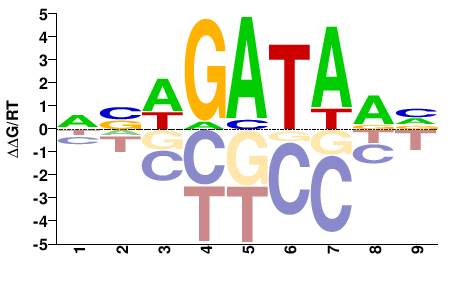 | Inferred - GATA4 (91% AA Identity, Homo sapiens) |
| Misc | HOMER | 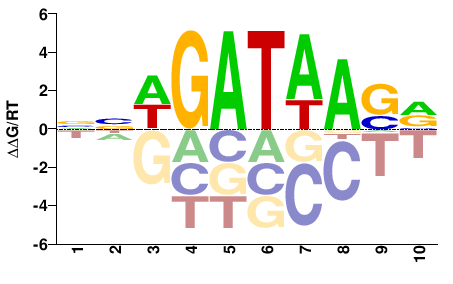 | Inferred - Gata4 (91% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 |  | Inferred - GATA6 (87% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - GATA6 (87% AA Identity, Homo sapiens) |
| PBM | Badis09 |  | Inferred - Gata6 (87% AA Identity, Mus musculus) |
| HT-SELEX | Nitta2015 | 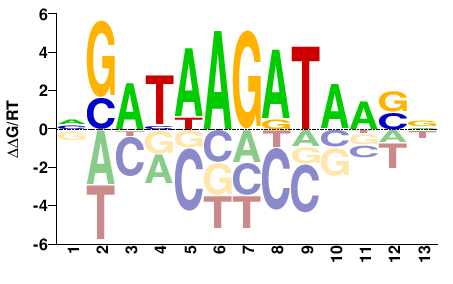 | Inferred - pnr (87% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 |  | Inferred - pnr (87% AA Identity, Drosophila melanogaster) |
| Transfac | Transfac | License required | Inferred - GATA6 (87% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA6 (87% AA Identity, Homo sapiens) |
| ChIP-chip | JASPAR |  | Inferred - pnr (87% AA Identity, Drosophila melanogaster) |
| ChIP-seq | modENCODE |  | Inferred - pnr (87% AA Identity, Drosophila melanogaster) |
| Misc | HocoMoco | 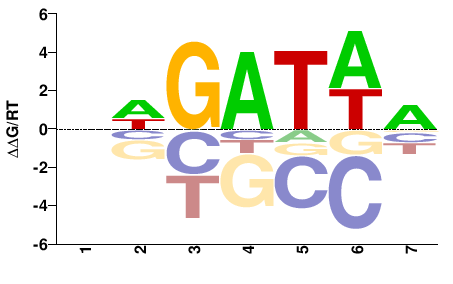 | Inferred - GATA6 (87% AA Identity, Homo sapiens) |
| HT-SELEX | Nitta2015 |  | Inferred - grn (84% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 | 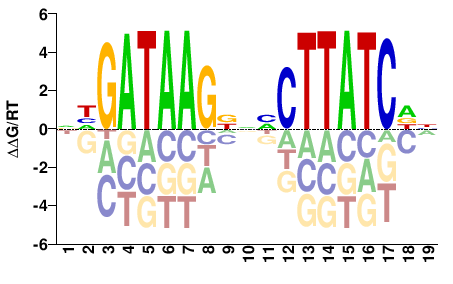 | Inferred - grn (84% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 | 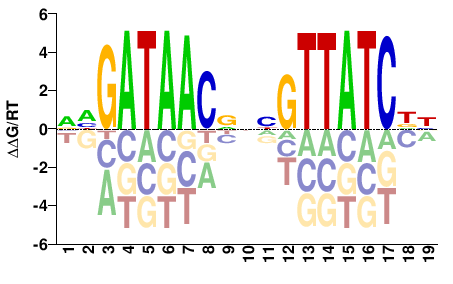 | Inferred - grn (84% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 | 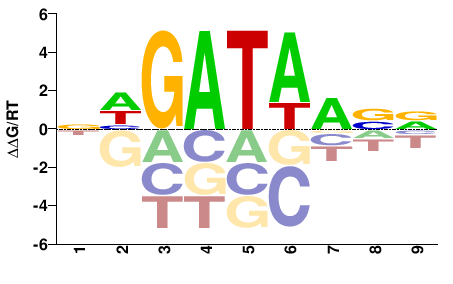 | Inferred - srp (84% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 |  | Inferred - srp (84% AA Identity, Drosophila melanogaster) |
| Misc | iDMMPMM |  | Inferred - srp (84% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Yin2017 |  | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 | 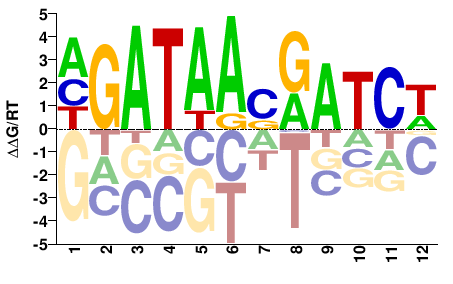 | Inferred - GATA3 (82% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - GATA3 (82% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - GATA3 (82% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - GATA3 (82% AA Identity, Homo sapiens) |
| PBM | Badis09 | 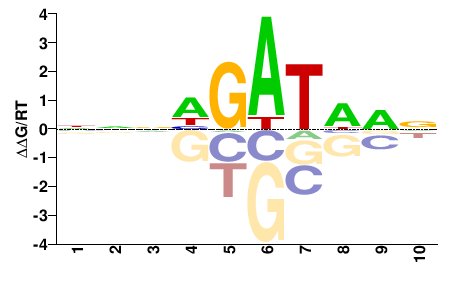 | Inferred - Gata3 (82% AA Identity, Mus musculus) |
| SMiLE-seq | Isakova2017 |  | Inferred - Gata3 (82% AA Identity, Mus musculus) |
| ChIP-seq | JASPAR | 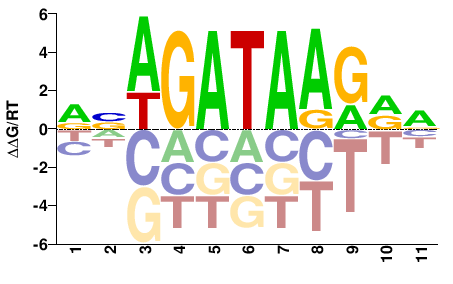 | Inferred - Gata1 (82% AA Identity, Mus musculus) |
| ChIP-seq | ENCODE |  | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| ChIP-seq | ENCODE | 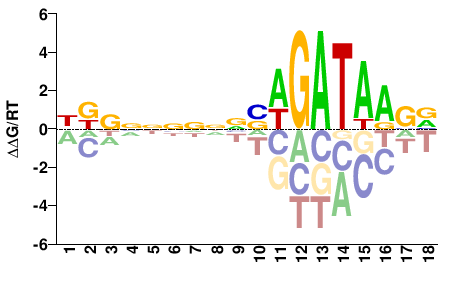 | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| ChIP-seq | ENCODE | 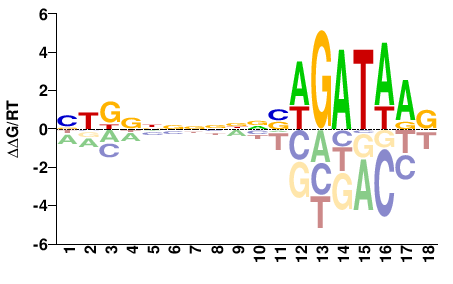 | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| ChIP-seq | ENCODE |  | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| ChIP-seq | ENCODE |  | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| ChIP-seq | JASPAR | 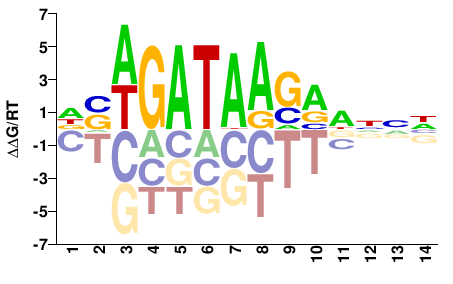 | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| ChIP-seq | ENCODE | 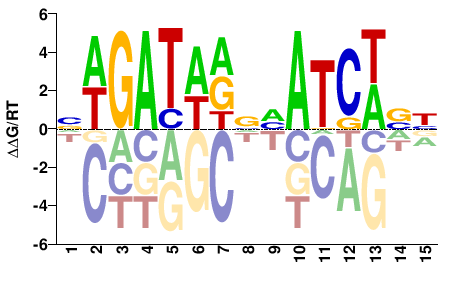 | Inferred - GATA3 (82% AA Identity, Homo sapiens) |
| ChIP-seq | JASPAR |  | Inferred - GATA3 (82% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA3 (82% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA3 (82% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA3 (82% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA3 (82% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA3 (82% AA Identity, Homo sapiens) |
| Misc | HocoMoco |  | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| Misc | HOMER |  | Inferred - GATA2 (82% AA Identity, Homo sapiens) |
| Misc | HocoMoco | 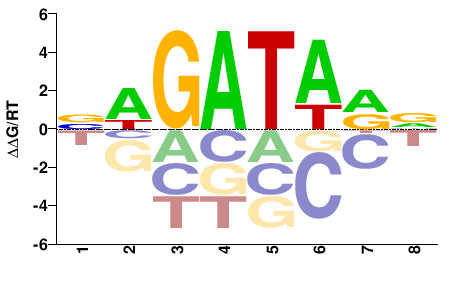 | Inferred - GATA3 (82% AA Identity, Homo sapiens) |
| Misc | HOMER | 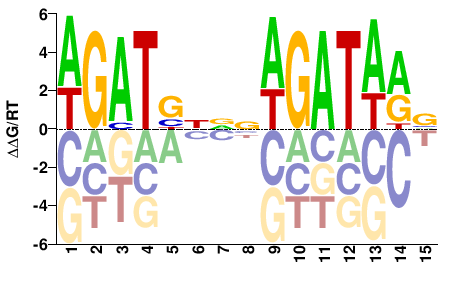 | Inferred - Gata3 (82% AA Identity, Mus musculus) |
| Misc | HOMER | 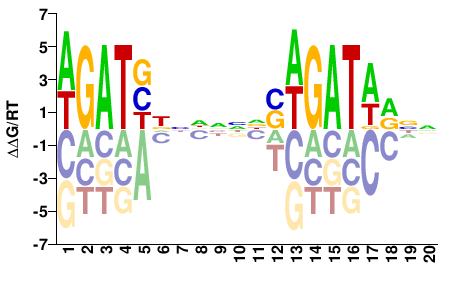 | Inferred - Gata3 (82% AA Identity, Mus musculus) |
| Misc | HOMER | 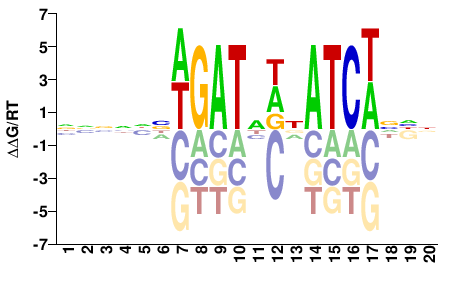 | Inferred - Gata3 (82% AA Identity, Mus musculus) |
| Misc | HOMER | 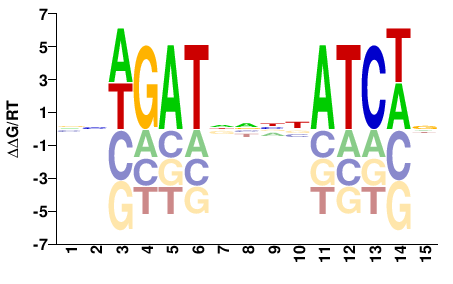 | Inferred - Gata3 (82% AA Identity, Mus musculus) |
| Misc | HOMER | 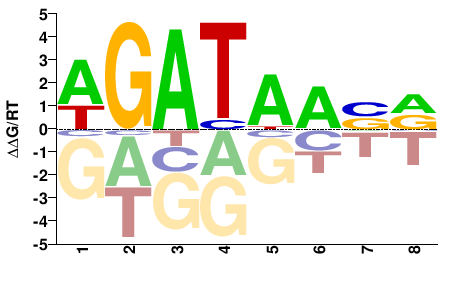 | Inferred - Gata3 (82% AA Identity, Mus musculus) |
| HT-SELEX | Jolma2010 | 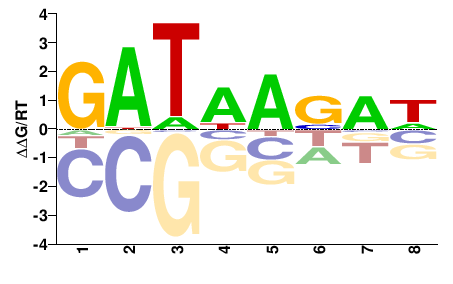 | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| HT-SELEX | Jolma2010 |  | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 | 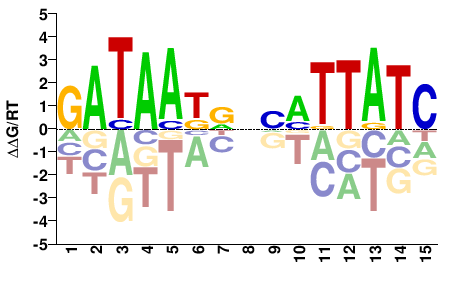 | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 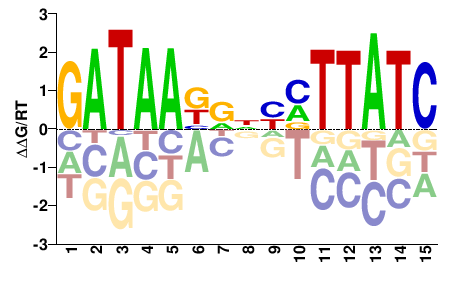 | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| ChIP-seq | ENCODE |  | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| ChIP-seq | ENCODE | 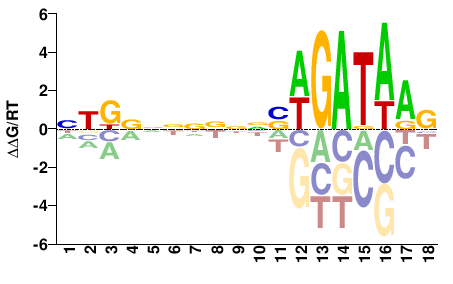 | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| ChIP-seq | modENCODE | 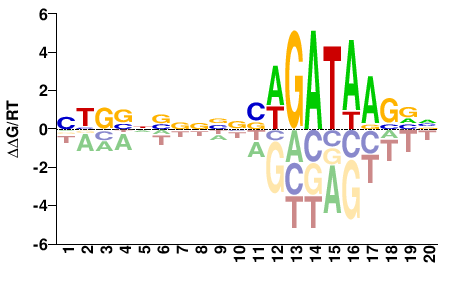 | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| ChIP-seq | modENCODE | 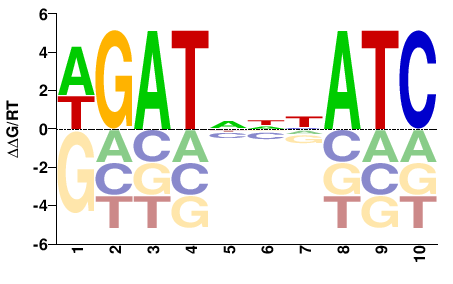 | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| Misc | HocoMoco |  | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| Misc | HOMER | 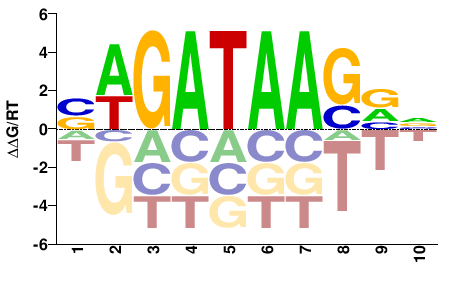 | Inferred - GATA1 (81% AA Identity, Homo sapiens) |
| ChIP-seq | modENCODE | 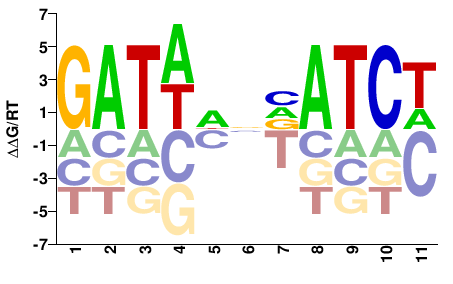 | Inferred - elt-1 (74% AA Identity, Caenorhabditis elegans) |
| ChIP-seq | modENCODE |  | Inferred - elt-1 (74% AA Identity, Caenorhabditis elegans) |