| Source |
Annotation |
Motif |
Evidence |
| HT-SELEX | Jolma2013 |  | Direct |
| HT-SELEX | Yin2017 |  | Direct |
| Methyl-HT-SELEX | Yin2017 | 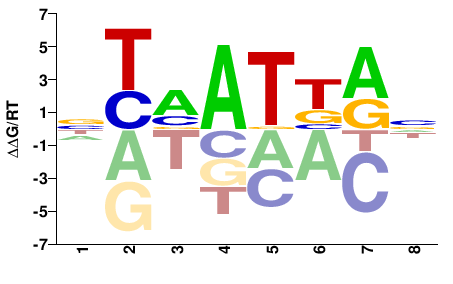 | Direct |
| Methyl-HT-SELEX | Yin2017 | 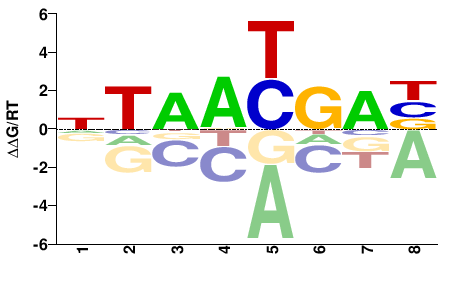 | Direct |
| Misc | HocoMoco |  | Direct |
| PBM | Berger08 | 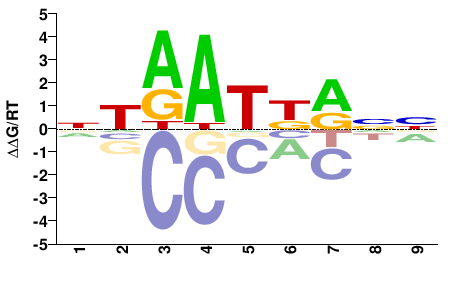 | Inferred - Hoxa1 (100% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 |  | Inferred - HOXB1 (87% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - HOXB1 (87% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - HOXB1 (87% AA Identity, Homo sapiens) |
| PBM | Berger08 | 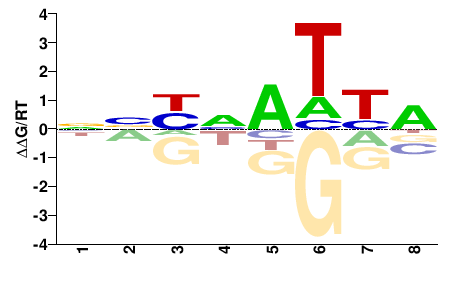 | Inferred - Hoxd1 (87% AA Identity, Mus musculus) |
| B1H | JASPAR |  | Inferred - lab (87% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 | 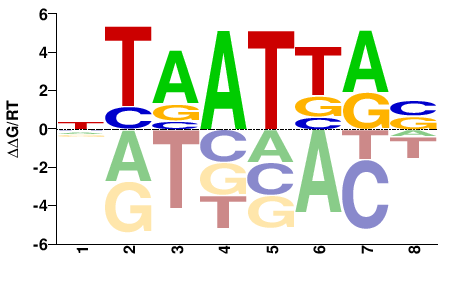 | Inferred - lab (87% AA Identity, Drosophila melanogaster) |
| PBM | Zoo_01 | 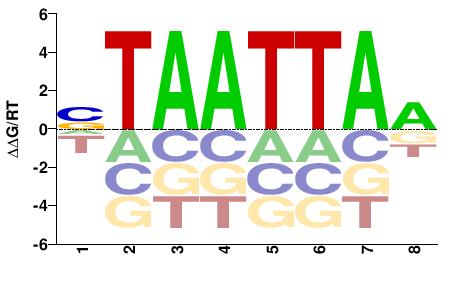 | Inferred - O76842_CUPSA (87% AA Identity, Cupiennius salei) |
| Misc | HocoMoco |  | Inferred - HOXB1 (87% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - HOXD1 (85% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 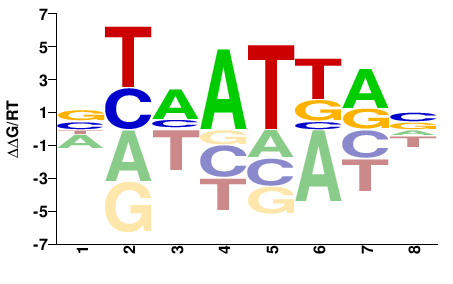 | Inferred - HOXD1 (85% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 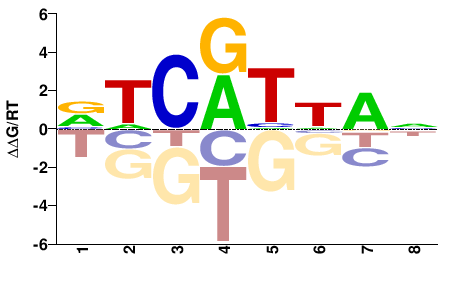 | Inferred - HOXD1 (85% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - HOXA2 (70% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 | 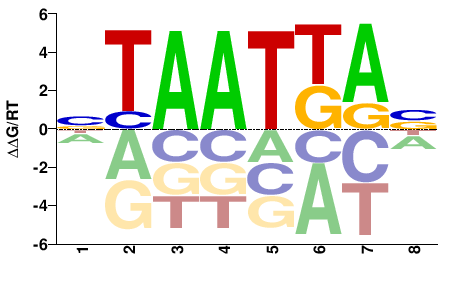 | Inferred - HOXA2 (70% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - HOXA2 (70% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 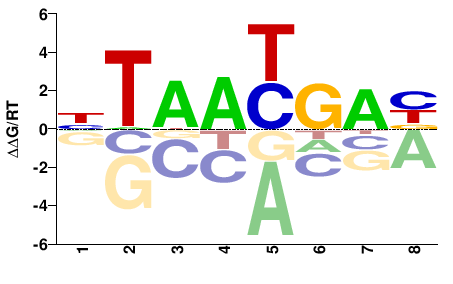 | Inferred - HOXA2 (70% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - HOXA2 (70% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 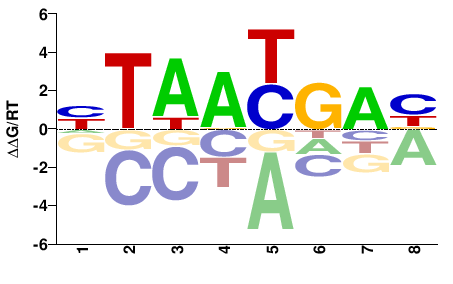 | Inferred - HOXA2 (70% AA Identity, Homo sapiens) |
| PBM | Zoo_01 | 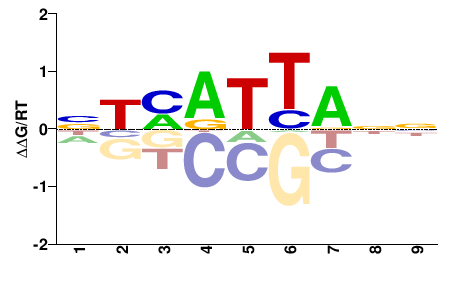 | Inferred - HOXA2 (70% AA Identity, Homo sapiens) |
| PBM | Berger08 | 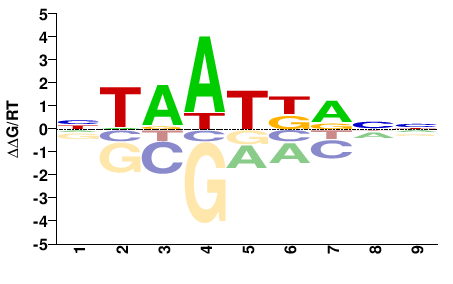 | Inferred - Hoxa2 (70% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 |  | Inferred - HOXB2 (70% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - HOXB2 (70% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 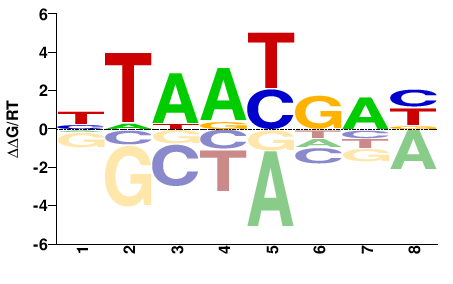 | Inferred - HOXB2 (70% AA Identity, Homo sapiens) |
| B1H | JASPAR |  | Inferred - pb (70% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 | 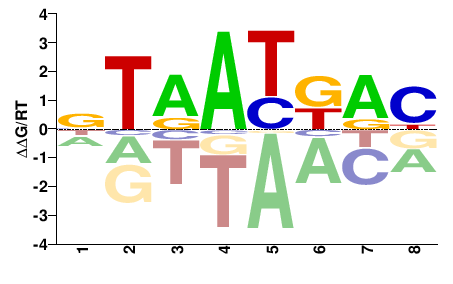 | Inferred - pb (70% AA Identity, Drosophila melanogaster) |
| PBM | Zoo_01 |  | Inferred - Q24782_9CNID (70% AA Identity, Eleutheria dichotoma) |
| PBM | Zoo_01 |  | Inferred - Q7M3U4_ACRFO (70% AA Identity, Acropora formosa) |
| Misc | HOMER |  | Inferred - Hoxa2 (70% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 |  | Inferred - HOXA4 (68% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 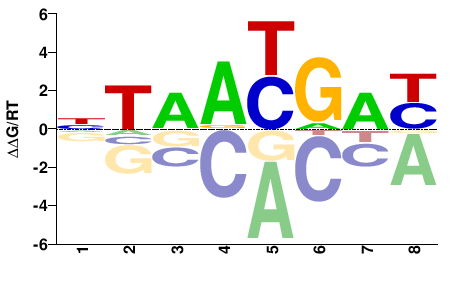 | Inferred - HOXA4 (68% AA Identity, Homo sapiens) |
| PBM | Berger08 | 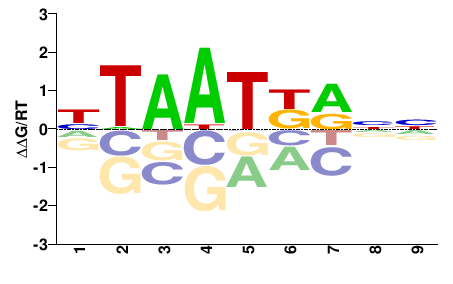 | Inferred - Hoxa4 (68% AA Identity, Mus musculus) |
| PBM | Berger08 | 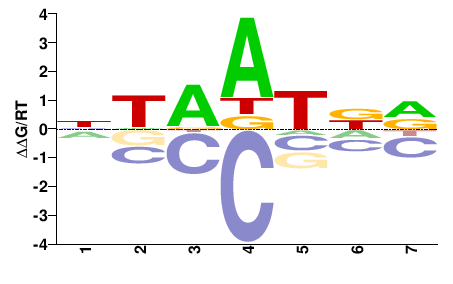 | Inferred - Hoxc5 (68% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 |  | Inferred - HOXD3 (68% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - HOXD3 (68% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - HOXD3 (68% AA Identity, Homo sapiens) |
| PBM | Berger08 | 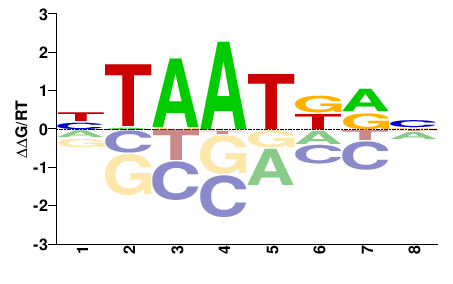 | Inferred - Hoxd3 (68% AA Identity, Mus musculus) |
| Transfac | Transfac | License required | Inferred - HOXA4 (68% AA Identity, Homo sapiens) |
| PBM | Badis09 | 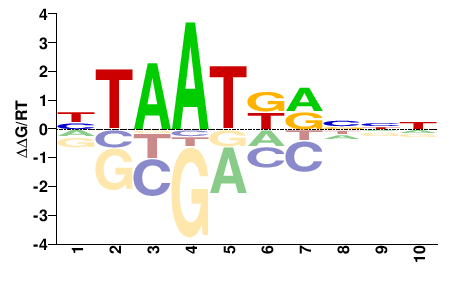 | Inferred - Hoxa3 (66% AA Identity, Mus musculus) |
| PBM | Berger08 | 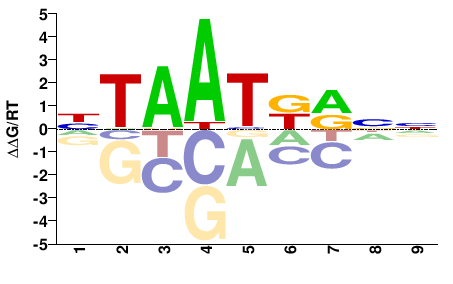 | Inferred - Hoxa3 (66% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 |  | Inferred - HOXA5 (66% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - HOXA5 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - HOXA5 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 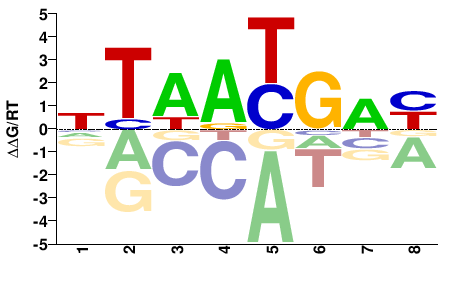 | Inferred - HOXA5 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - HOXA5 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 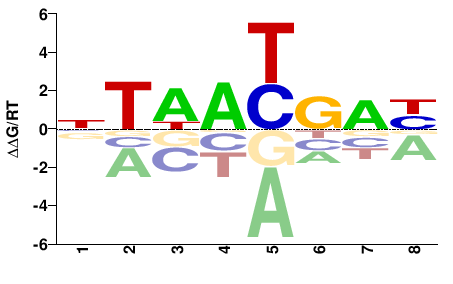 | Inferred - HOXA5 (66% AA Identity, Homo sapiens) |
| PBM | Berger08 |  | Inferred - Hoxa5 (66% AA Identity, Mus musculus) |
| PBM | Berger08 | 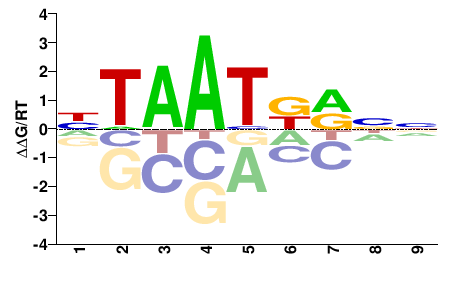 | Inferred - Hoxb3 (66% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 |  | Inferred - HOXB4 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 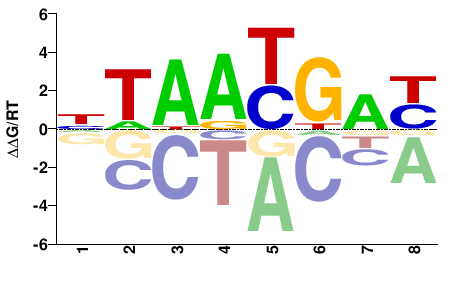 | Inferred - HOXB4 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 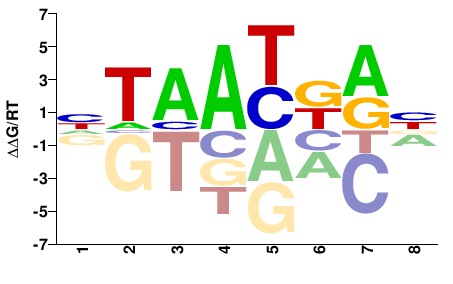 | Inferred - HOXB4 (66% AA Identity, Homo sapiens) |
| PBM | Berger08 |  | Inferred - Hoxb4 (66% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 |  | Inferred - HOXB5 (66% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - HOXB5 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 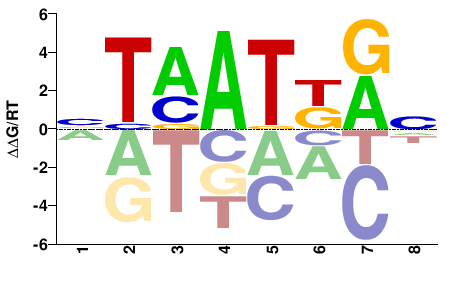 | Inferred - HOXB5 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 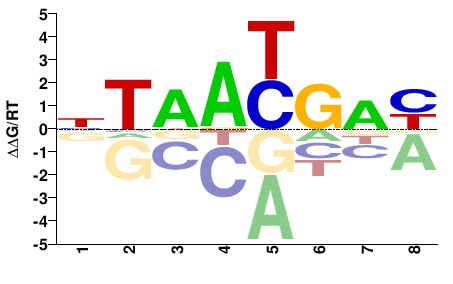 | Inferred - HOXB5 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - HOXB5 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 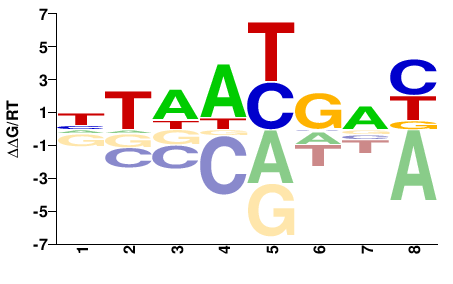 | Inferred - HOXB5 (66% AA Identity, Homo sapiens) |
| PBM | Berger08 |  | Inferred - Hoxb5 (66% AA Identity, Mus musculus) |
| PBM | Barrera2016 |  | Inferred - HOXC4 (66% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 |  | Inferred - HOXC4 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - HOXC4 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 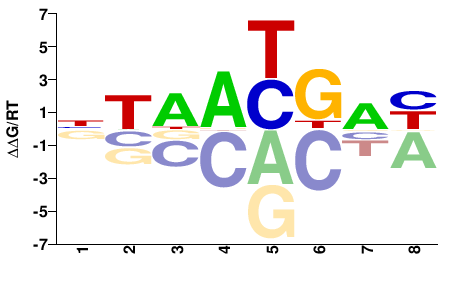 | Inferred - HOXC4 (66% AA Identity, Homo sapiens) |
| PBM | Berger08 | 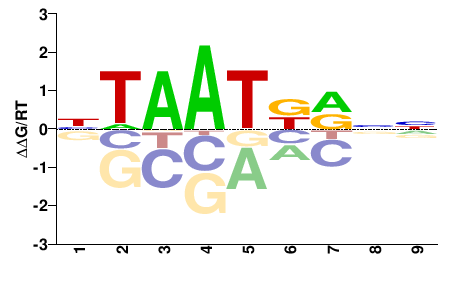 | Inferred - Hoxc4 (66% AA Identity, Mus musculus) |
| HT-SELEX | Yin2017 |  | Inferred - HOXD4 (66% AA Identity, Homo sapiens) |
| HT-SELEX | Yin2017 | 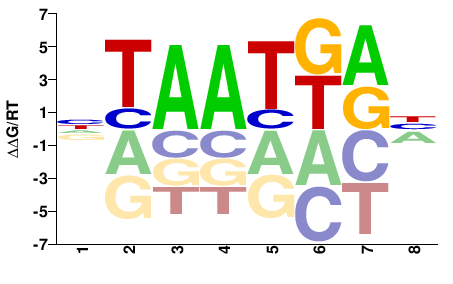 | Inferred - HOXD4 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 |  | Inferred - HOXD4 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 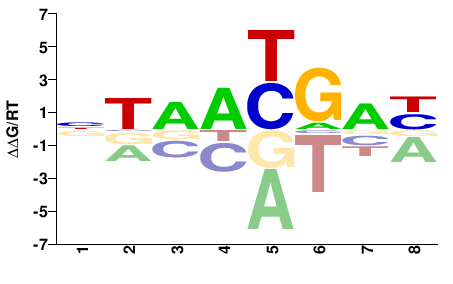 | Inferred - HOXD4 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 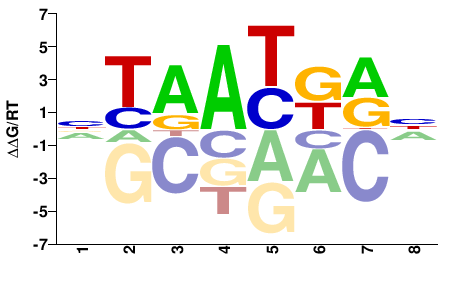 | Inferred - HOXD4 (66% AA Identity, Homo sapiens) |
| Methyl-HT-SELEX | Yin2017 | 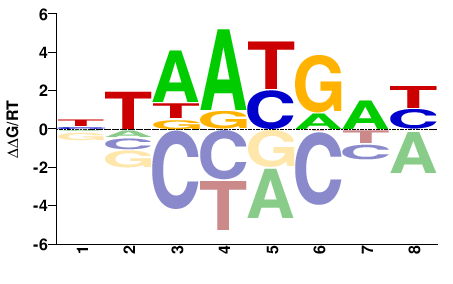 | Inferred - HOXD4 (66% AA Identity, Homo sapiens) |
| PBM | Zoo_01 |  | Inferred - Q0N4M8_NEMVE (66% AA Identity, Nematostella vectensis) |
| B1H | JASPAR |  | Inferred - Scr (66% AA Identity, Drosophila melanogaster) |
| HT-SELEX | Nitta2015 | 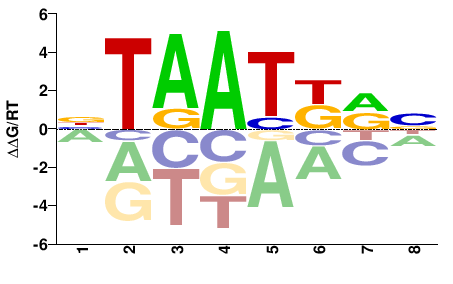 | Inferred - Scr (66% AA Identity, Drosophila melanogaster) |
| Transfac | Transfac | License required | Inferred - HOXA3 (66% AA Identity, Homo sapiens) |
| COMPILED | JASPAR | 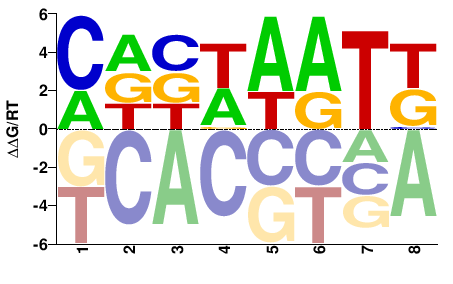 | Inferred - HOXA5 (66% AA Identity, Homo sapiens) |
| Transfac | Transfac | License required | Inferred - HOXA5 (66% AA Identity, Homo sapiens) |
| ChIP-seq | modENCODE | 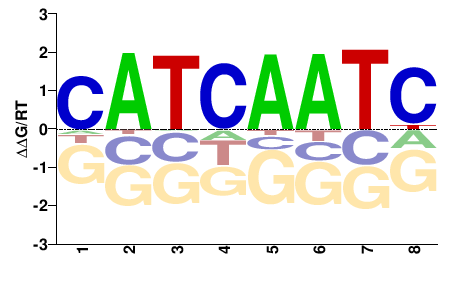 | Inferred - lin-39 (66% AA Identity, Caenorhabditis elegans) |
| Misc | HocoMoco |  | Inferred - HOXA5 (66% AA Identity, Homo sapiens) |
| Misc | HOMER | 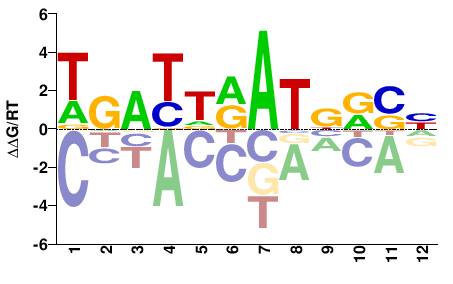 | Inferred - Hoxb4 (66% AA Identity, Mus musculus) |
| Misc | HocoMoco | 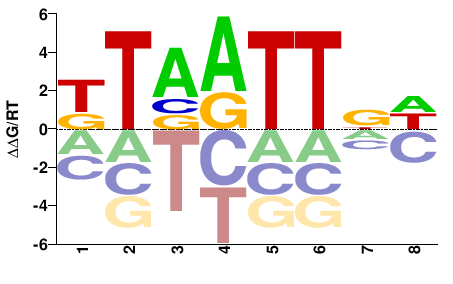 | Inferred - HOXD4 (66% AA Identity, Homo sapiens) |
| Misc | HOMER |  | Inferred - lin-39 (66% AA Identity, Caenorhabditis elegans) |