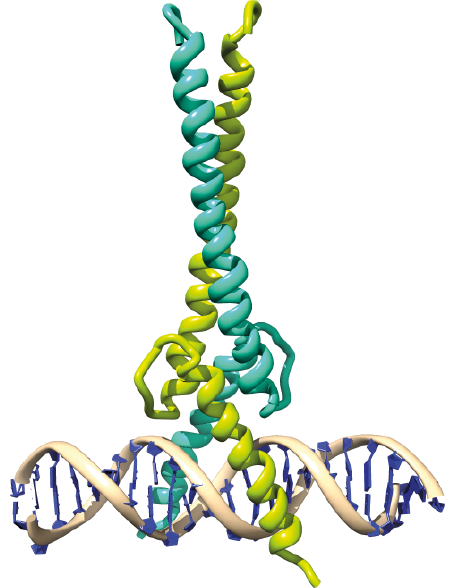
|
| AHR |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
In vivo/Misc source |
Only known motifs are from Transfac or HocoMoco - origin is uncertain |
|
|
| AHRR |
bHLH |
Yes |
Inferred motif |
1 Monomer or homomultimer |
In vivo/Misc source |
|
|
|
| ARNT |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
100 perc ID - in vitro |
|
|
|
| ARNT2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| ARNTL |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| ARNTL2 |
bHLH |
Yes |
Inferred motif |
2 Obligate heteromer |
High-throughput in vitro |
|
ARNTL2 Forms heterodimers with CLOCK and NPAS2 (PMID:10864977 ). |
|
| ASCL1 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| ASCL2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| ASCL3 |
bHLH |
Yes |
Inferred motif |
2 Obligate heteromer |
High-throughput in vitro |
|
ASCL1 heterodimerizes with TCF3; TCF4 and TCF12 (PMID: 24835951). |
|
| ASCL4 |
bHLH |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
Probably heterodimerizes with TCF3; TCF4 or TCF12 like the other ASCL TFs |
|
| ASCL5 |
bHLH |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
Probably heterodimerizes with TCF3; TCF4 or TCF12 like the other ASCL TFs |
|
| ATOH1 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| ATOH7 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| ATOH8 |
bHLH |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| BHLHA15 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| BHLHA9 |
bHLH |
Yes |
Likely to be sequence specific TF |
2 Obligate heteromer |
No motif |
|
Protein heterodimerizes with TCF3; TCF4 and TCF12 (PMID: 25466284). |
|
| BHLHE22 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| BHLHE23 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| BHLHE40 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| BHLHE41 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| CCDC169-SOHLH2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
100 perc ID - in vitro |
|
|
|
| CLOCK |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
Can also bind as heterodimer with ARNTL. |
|
| EPAS1 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
In vivo/Misc source |
Only known motifs are from Transfac or HocoMoco - origin is uncertain |
|
|
| FERD3L |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| FIGLA |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| HAND1 |
bHLH |
Yes |
Known motif |
2 Obligate heteromer |
In vivo/Misc source |
Only known motifs are from Transfac or HocoMoco - origin is uncertain |
Obligate heteromer (PMID: 10611232). |
|
| HAND2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| HELT |
bHLH |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
Can form homodimers or heterodimers with HEY2 and HES5, suggesting also that the HELT homodimer is dependent of a additional Orange domain while the HEY2 heterodimer requires just the DBD (PMID: 14764602). |
|
| HES1 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| HES2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| HES3 |
bHLH |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| HES4 |
bHLH |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| HES5 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| HES6 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| HES7 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
Also forms heterodimers. |
|
| HEY1 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| HEY2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| HEYL |
bHLH |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| HIF1A |
bHLH |
Yes |
Known motif |
2 Obligate heteromer |
In vivo/Misc source |
Only known motifs are from Transfac or HocoMoco - origin is uncertain |
Binds as obligate heteromer with ARNT (PMID: 9027737). |
|
| HIF3A |
bHLH |
Yes |
Inferred motif |
2 Obligate heteromer |
In vivo/Misc source |
|
Binds DNA as a heterodimer with ARNT and ARNT2; reviewed in (PMID: 24099156). |
|
| ID1 |
bHLH |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
ID bHLH proteins lack the basic region and should not be able to bind DNA. The HT-SELEX motif for ID4 is likely by a co-precipitated protein or it is a contamination |
|
| ID2 |
bHLH |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
ID bHLH proteins lack the basic region and should not be able to bind DNA. The HT-SELEX motif for ID4 is likely by a co-precipitated protein or it is a contamination |
|
| ID3 |
bHLH |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
ID bHLH proteins lack the basic region and should not be able to bind DNA. The HT-SELEX motif for ID4 is likely by a co-precipitated protein or it is a contamination |
|
| ID4 |
bHLH |
No |
Unlikely to be sequence specific TF |
4 Not a DNA binding protein |
No motif |
|
ID bHLH proteins lack the basic region and should not be able to bind DNA. The HT-SELEX motif for ID4 is likely by a co-precipitated protein or it is a contamination |
|
| LYL1 |
bHLH |
Yes |
Inferred motif |
2 Obligate heteromer |
In vivo/Misc source |
|
TAL1, TAL2 and LYL are very similar and have a basic region with a bulky tryptophan inserting into the cluster of the basic residues. Binds DNA as a heterodimer with TCF3 (PDB:2YPB and PDB:2YPA). |
|
| MAX |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| MESP1 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| MESP2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| MITF |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
100 perc ID - in vitro |
|
Binds DNA both as homo- and heterodimers (PMID: 23207919). The structure 4ATI is a homodimer with DNA GTTAGCACATGACCCT. |
|
| MLX |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| MLXIP |
bHLH |
Yes |
Inferred motif |
2 Obligate heteromer |
High-throughput in vitro |
|
|
|
| MLXIPL |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| MNT |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
Prefers to heterodimerize with MAX over homodimer formation (PMID: 9000049). |
|
| MSC |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| MSGN1 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| MXD1 |
bHLH |
Yes |
Inferred motif |
2 Obligate heteromer |
In vivo/Misc source |
|
All three MXD proteins have very similar sequences and should behave accordingly, making heterodimers with at least MAX (PMID:8521822). |
|
| MXD3 |
bHLH |
Yes |
Inferred motif |
2 Obligate heteromer |
In vivo/Misc source |
|
All three MXD proteins have very similar sequences and should behave accordingly, making heterodimers with at least MAX (PMID:8521822). |
|
| MXD4 |
bHLH |
Yes |
Inferred motif |
2 Obligate heteromer |
In vivo/Misc source |
|
All three MXD proteins have very similar sequences and should behave accordingly, making heterodimers with at least MAX (PMID:8521822). |
|
| MXI1 |
bHLH |
Yes |
Known motif |
2 Obligate heteromer |
In vivo/Misc source |
Has a putative AT-hook |
Obligate heteromer (PMID: 8425219). |
|
| MYC |
bHLH |
Yes |
Known motif |
2 Obligate heteromer |
In vivo/Misc source |
|
Functions as a heterodimer with MAX. |
|
| MYCL |
bHLH |
Yes |
Inferred motif |
2 Obligate heteromer |
In vivo/Misc source |
|
Similar to MYC and thus likely to functions as a heterodimer with MAX. |
|
| MYCN |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| MYF5 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
In vivo/Misc source |
Only known motifs are from Transfac or HocoMoco - origin is uncertain |
|
|
| MYF6 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| MYOD1 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| MYOG |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| NCOA1 |
bHLH |
Yes |
Likely to be sequence specific TF |
2 Obligate heteromer |
No motif |
|
NCOA1/2/3 all have been tested on PBMs and HT-SELEX without yielding a motif. All are co-activators, collectively suggesting they might be obligate heteromers. |
|
| NCOA2 |
bHLH |
Yes |
Likely to be sequence specific TF |
2 Obligate heteromer |
No motif |
|
NCOA1/2/3 all have been tested on PBMs and HT-SELEX without yielding a motif. All are co-activators, collectively suggesting they might be obligate heteromers. |
|
| NCOA3 |
bHLH |
Yes |
Likely to be sequence specific TF |
2 Obligate heteromer |
No motif |
|
NCOA1/2/3 all have been tested on PBMs and HT-SELEX without yielding a motif. All are co-activators, collectively suggesting they might be obligate heteromers. |
|
| NEUROD1 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| NEUROD2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| NEUROD4 |
bHLH |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| NEUROD6 |
bHLH |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| NEUROG1 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| NEUROG2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| NEUROG3 |
bHLH |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| NHLH1 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| NHLH2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| NPAS1 |
bHLH |
Yes |
Inferred motif |
2 Obligate heteromer |
In vivo/Misc source |
|
Likely obligate heteromer (PMID: 27782878). |
|
| NPAS2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| NPAS3 |
bHLH |
Yes |
Inferred motif |
2 Obligate heteromer |
In vivo/Misc source |
|
Likely obligate heteromer (PMID: 9374395; PMID: 27782878). |
|
| NPAS4 |
bHLH |
Yes |
Inferred motif |
2 Obligate heteromer |
High-throughput in vitro |
|
Likely dimerizes with ARNT2 (PMID:24263188). |
|
| OLIG1 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| OLIG2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
OLIG2 can homodimerize and heterodimerize with OLIG1 and NGN2 (PMID:15655114; PMID: 21382552). |
|
| OLIG3 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| PTF1A |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| SCX |
bHLH |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
SCX heterodimerizes with TCF3 (PMID: 19828133). The Taipale lab has an unpublished HT-SELEX model; both it and the inferred B1H motif seem very weak, supporting the idea that the homodimer does not bind DNA well. |
|
| SIM1 |
bHLH |
Yes |
Inferred motif |
2 Obligate heteromer |
In vivo/Misc source |
|
Similar to SIM2, which is a likely obligate heteromer. |
|
| SIM2 |
bHLH |
Yes |
Inferred motif |
2 Obligate heteromer |
In vivo/Misc source |
|
Likely obligate hetereomer (PMID: 7592839) |
|
| SOHLH1 |
bHLH |
Yes |
Likely to be sequence specific TF |
2 Obligate heteromer |
No motif |
|
Forms SOHLH1-SOHLH2 heterodimers (PMID: 22056784). |
|
| SOHLH2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| SREBF1 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| SREBF2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| TAL1 |
bHLH |
Yes |
Known motif |
2 Obligate heteromer |
In vivo/Misc source |
Only known motifs are from Transfac or HocoMoco - origin is uncertain |
No motif yielded from PBMs or HT-SELEX. Binds DNA as heterodimer with TCF3 (PDB:2YPB and PDB:2YPA). |
|
| TAL2 |
bHLH |
Yes |
Inferred motif |
2 Obligate heteromer |
In vivo/Misc source |
|
No motif yielded from PBMs or HT-SELEX. Similar to TAL1, which binds DNA as heterodimer with TCF3 (PDB:2YPB and PDB:2YPA). |
|
| TCF12 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| TCF15 |
bHLH |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| TCF21 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| TCF23 |
bHLH |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
Basic region is very similar to TCF21, which binds DNA as a homodimer in HT-SELEX. |
|
| TCF24 |
bHLH |
Yes |
Inferred motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| TCF3 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| TCF4 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| TCFL5 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| TFAP4 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
Also can form heterodimers (PMID: 26550823). |
|
| TFE3 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| TFEB |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| TFEC |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| TWIST1 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
In vivo/Misc source |
Only known motifs are from Transfac or HocoMoco - origin is uncertain |
Can form both homodimers and heterodimers with TCF3 (PMID: 16502419). |
|
| TWIST2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
100 perc ID - in vitro |
|
|
|
| USF1 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
|
|
|
| USF2 |
bHLH |
Yes |
Known motif |
1 Monomer or homomultimer |
High-throughput in vitro |
Has a putative AT-hook |
|
|
| USF3 |
bHLH |
Yes |
Likely to be sequence specific TF |
1 Monomer or homomultimer |
No motif |
|
|